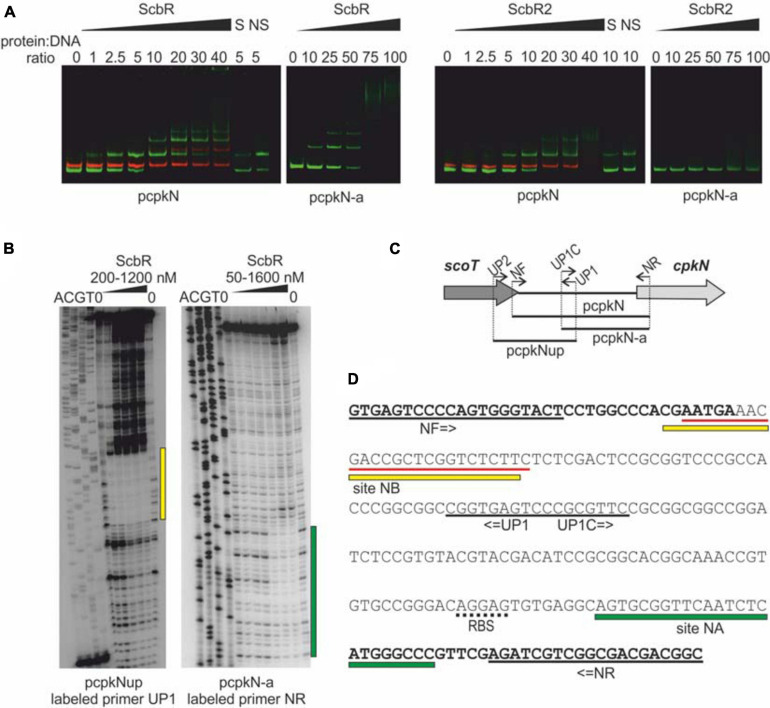
FIGURE 5.
Binding of His-tagged ScbR and ScbR2 proteins to cpkN promoter region. (A) Electrophoretic mobility shift assay. Green bands – IRDye 800 labeled fragments pcpkN and pcpkN-a, red bands – IRDye 700 labeled negative control fragment MYCO, S, 10-fold excess of unlabeled specific competitor DNA (pcpkN); NS, 10-fold excess of unlabeled non-specific competitor DNA (MYCO). (B) Protection of cpkN promoter region from digestion by ScbR protein (DNase I footprint). The amplified fragments and the radiolabeled primers are indicated below the lanes. Protected regions are marked with yellow and green bars. (C) Schematic representation of the location of primers and fragments used for EMSA and footprint experiments. (D) Nucleotide sequence of pcpkN fragment. Coding sequences of scoT and cpkN genes are in bold. Predicted ribosome binding site (RBS) is underlined with a dotted line. Primer sequences are underlined with black solid lines. Regions protected by ScbR protein from (C) are marked with yellow and green bars. Sequence recognized by ScbR2 reported by Li et al. (2015) is underlined with red lines.