Figure 1.
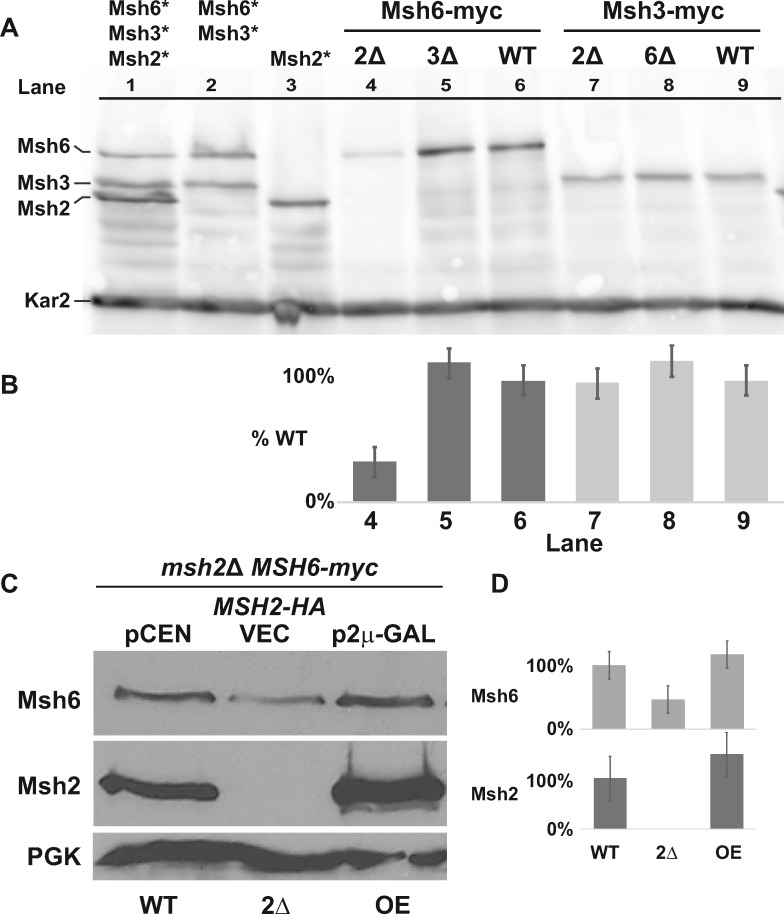
Levels of Msh6, but not Msh3, are influenced by Msh2. (A) Chromosomally tagged Msh6 and Msh3 proteins display differential regulation in the absence of Msh2. Cultures grown to mid-exponential phase were processed and immunoblotted with α-myc antibodies for Msh2-myc, Msh3-myc, and Msh6-myc, and α-Kar2 for the loading control. Lanes 1-3 show the relative migration position and levels of Msh6, Msh3, or Msh2 expressed from stains where the proteins are identically tagged with the myc epitope, singly or in combinations (lanes 1-3, tagged protein indicated above the lanes with asterisks). Msh6-myc (lanes 4-6) and Msh3-myc (lanes 7-8) proteins were expressed in the absence of Msh2 (2Δ), Msh3 (3Δ) or Msh6 (6Δ), or in wild-type (WT) strain backgrounds. (B) Band intensities from Panel A of Msh6-myc and Msh3-myc were normalized to the loading controls using ImageJ and graphed as the percentage of each protein expressed in the WT strain (%WT). Lane numbers from Panel A are shown for reference. Error bars represent the stand error. (C) Msh6 levels are controlled by the abundance of Msh2. A strain with a chromosomally myc tagged MSH6 (MSH6-myc) and with a deletion in MSH2 (msh2Δ MSH6-myc) harbored plasmids expressing wild-type MSH2-HA from the endogenous promoter on a low copy, centromere-based plasmid (pCEN or WT), or overexpressed from an inducible GAL10 promoter on a high-copy, 2 μ plasmid (p2μ-GAL or OE). As a comparison, no Msh2 was expressed in the msh2Δ MSH6-myc strain with a plasmid vector (VEC, or 2Δ). The cells were grown to exponential phase in 2% galactose. Msh6-myc, Msh2-HA, and the PGK loading control were detected by α-myc, α-HA, and α-PGK, respectively. (D) Band intensities from Panel C of Msh6-myc and Msh2-HA were normalized to the loading controls using ImageJ and graphed as the percentage of each protein expressed in the WT strain described in Panel C. Error bars represent the stand error.