Figure 4.
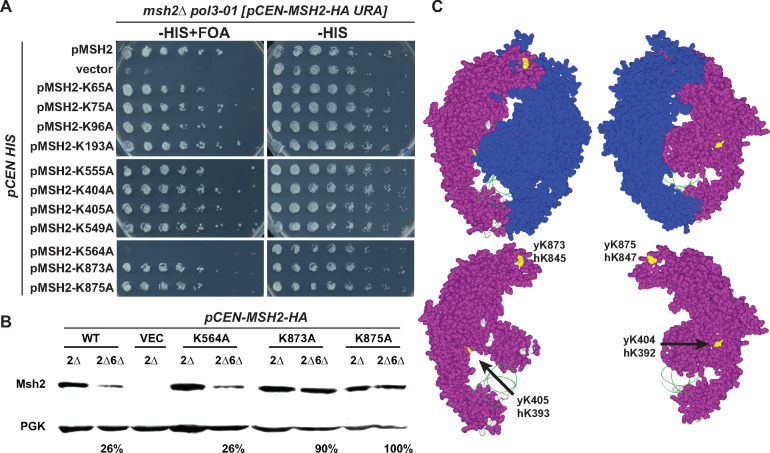
Conserved lysines on the MutSα stabilize Msh2 in the absence of Msh6. (A) Functional assays of lysine substitution mutants. An msh2Δ pol3-01 strain kept alive by a URA3-based plasmid expressing wild-type MSH2 (pCEN-MSH2-HA URA) was transformed with HIS3 encoding plasmids (pCEN HIS) expressing endogenously expressed wild-type MSH2 (pMSH2), no MSH2 (vector), and lysine substitution variants (pMSH2-KcodonA). Stains were grown overnight in medium lacking histidine allowing for the loss of the covering wild-type MSH2 URA3 plasmid. Fivefold serial dilutions were delivered to plates lacking histidine with no drug (–HIS) or supplement with 5-FOA (–HIS+ FOA). 5-FOA selects for cells that were able to lose the MSH2 URA3 plasmid during growth, indicating full suppression of the mismatch repair defect. (B) Steady-state levels of Msh2 and its lysine substitution variants in the presence (2Δ) and absence of Msh6 (2Δ6Δ). The proteins were detected as described in Figure 1A. Band intensities were normalized to the loading controls using ImageJ and shown below the immunoblot images as the percentage of the protein level expressed in the presence of Msh6. (C) Conserved lysines highlighted on the human MutSα structure. The Msh2 protein is shown in purple with the conserved lysines that when mutated had no effect on levels (gray) or stabilized the levels of Msh2 (yellow) in the absence of Msh6. The ribbon backbone of the DNA molecule (green) is shown for orientation purposes. Two views of the heterodimer with and without Msh6 (blue) are shown. Images created without Msh6 are to reveal the concealed lysines. The potential lysine targets (yellow) are labeled with the yeast and human codon numbers in the images with just Msh2. Images were made with Swiss PDB Viewer (Guex et al. 1999) and Persistence of Vision Raytracer (Version 3.6) retrieved from http://www.povray.org/download/.