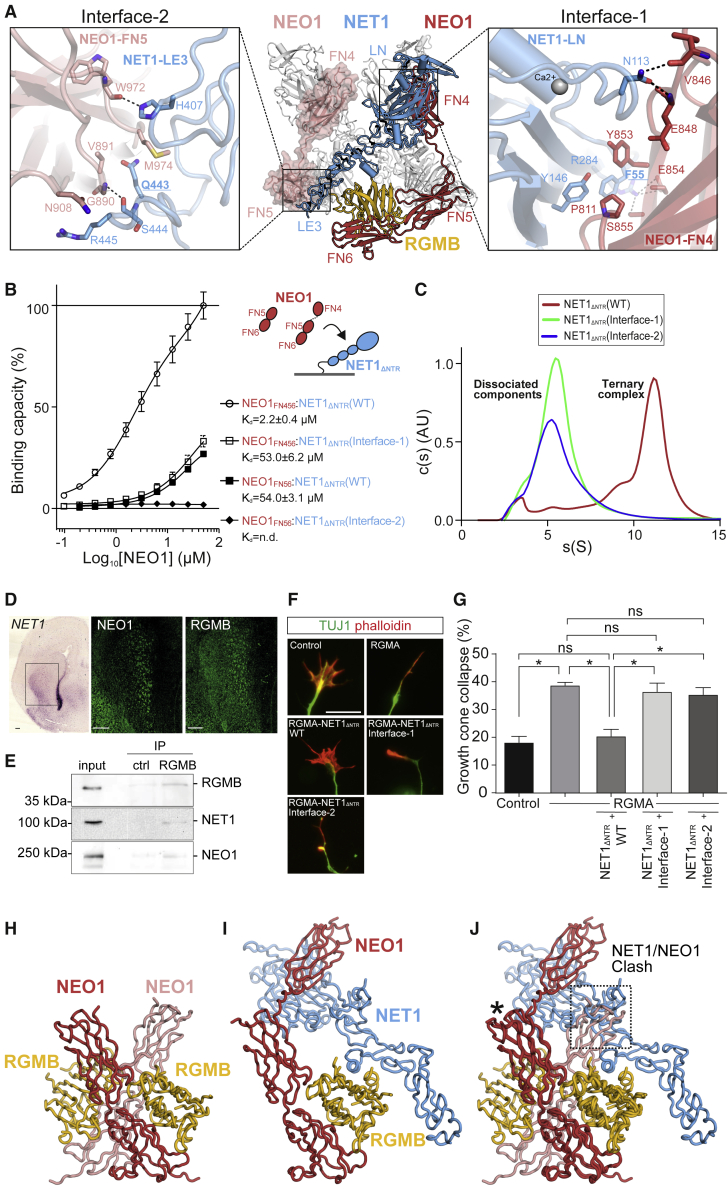
Figure 3.
Interface analysis of the ternary NEO1-NET1-RGMB super-complex
(A) Close-up views of the observed NET1-NEO1 interfaces (right: interface 1, left: interface 2). Residues are displayed in stick representation and labelled according to domain color-coding. A Ca2+ ion bound to NET1 LN (grey sphere) and hydrogen bonds (dashed black lines) are displayed. Mutated residues are in bold and underlined.
(B) SPR equilibrium binding curves for the NET1-NEO1 interaction. A schematic of the experiment and the calculated Kd values are shown.
(C) AUC analysis of the NEO1FN456-NET1ΔNTR-RGMBECD complex, using NET1ΔNTR WT and mutants. Both NET1 interface-1 and -2 mutants abolish the 3:3:3 stoichiometry of the NEO1-NET1-RGMB super-complex.
(D) Overlapping expression of NET1 RNA (in situ hybridization), and NEO1 and RGMB protein (immunohistochemistry) in consecutive coronal sections of E16 mouse striatum. Boxed area is shown at higher magnification for NEO1 and RGMB. Scale bar, 100 μm.
(E) RGMB immunoprecipitation (IP) from adult mouse cortex was followed by immunoblotting. Input samples (lane 1), IP using control non-specific IgGs (cntrl) (lane 2), and anti-RGMB IP (lane 3). NEO1 and NET1 co-IP with RGMB from adult mouse brain lysates.
(F and G) Functional analysis of the effect of NET1 on RGMA-mediated growth cone collapse.
(F) Representative examples of growth cones from mouse P0 cortical neurons. Neurons were stained with the microtubule marker Tuj1 (green) and F-actin marker phalloidin (red). Scale bar, 10 μm.
(G) Quantification of growth cone collapse. Growth cones were treated with control or RGMA alone and in combination with different NET1 variants. Proportions of collapsed growth cones relative to control are displayed. n = 3 experiments, one-way ANOVA followed by Tukey’s multiple comparison test. ∗p < 0.05. Data are shown as means ± SEM.
(H–J) Comparison of binary NEO1-RGM (PDB ID 4BQ6 [Bell et al., 2013]) and the ternary NEO1-NET1-RGMB complexes shown as ribbons. The ternary NEO1-NET1-RGMB protomer complex architecture (I) clashes with the NEO1-RGM dimer-of-dimers signaling conformation (H) when superimposed on NEO1 (marked with an asterisk) (J). See also Figure S3, Figure S4, Figure S5.