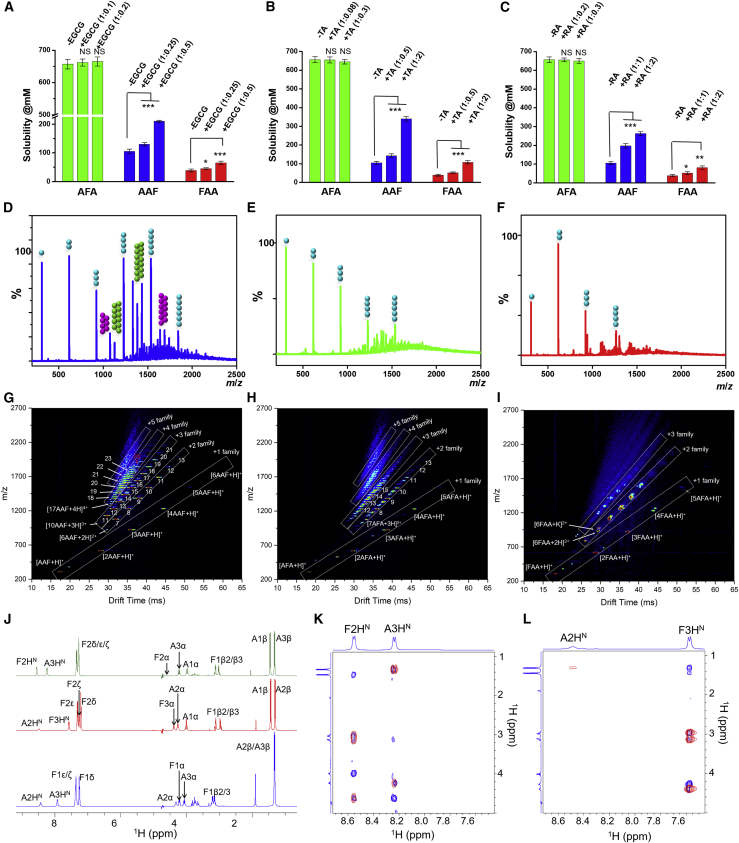
Figure 3.
Structure-dependent solubility of the tripeptides
(A–C) The effect of the polyphenols EGCG (A), TA (B), and RA (C) on modulation of solubility of the tripeptides because of interference with structural organization. Statistical distribution and p values of tripeptides solubility by the inhibitors compared with their absence is shown. The data represent the mean ± SEM for 3 independent experiments. N.S., not significant; ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001.
(D–F) ESI-MS mass spectra of (D) AAF, (E) AFA, and (F) FAA (10 mM in water, pH 6.8, 25°C). Circles above the peaks denote the oligomer order. The number of circles represents the number of monomers in the aggregate. +1, +2, and +3 charge states are represented as cyan, pink, and green circles, respectively.
(G–I) ESI-IMS-MS 2D plot of the (G) AAF, (H) AFA, and (I) FAA monomers through to oligomers. ESI-IMS-MS 2D plots show the IMS drift time versus mass/charge (m/z) versus intensity (z, square-root scale). The different charge families are marked with white boxes. The numbers indicate the corresponding oligomeric states (for details, see Supplemental information).
(J) One-dimensional 1H NMR spectra of tripeptides (FAA, blue; AAF, red; AFA, green) dissolved in 90% H2O/10% 2H2O, with assignments labeled.
(K and L) 2D [1H-1H]-NOESY (blue) and [1H-1H]-TOCSY (red) NMR spectra of AFA (K) and AAF (L). For clarity, only the fingerprint region of TOCSY/NOESY spectra is shown, and the assignments are labeled.