Abstract
Liver cancer is one of the most common malignant human tumors with the highest morbidity and mortality rates of all cancer types in China. Evidence suggests that long non-coding RNA prostate cancer-associated transcript 6 (PCAT6) plays an essential role in tumor progression. However, the roles and mechanism of PCAT6 in liver cancer remain unclear. The present study showed that the expression of PCAT6 and heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) was upregulated in liver cancer tissues compared with non-cancerous tissues and were associated with poor overall survival time, whereas microRNA (miR)-326 expression was downregulated. Moreover, knockdown of PCAT6 significantly inhibited the proliferation and invasion of liver cancer cells in vitro and in vivo. A dual-luciferase reporter gene assay demonstrated that PCAT6 could bind to miR-326 and that hnRNPA2B1 was a direct target gene of miR-326. Mechanistically, silenced PCAT6 suppressed the malignant phenotype of liver cancer cells through upregulating the inhibitory effect of miR-326 on hnRNPA2B1 expression. Taken together, these data demonstrated that knockdown of PCAT6 inhibited liver cancer progression through regulation of the miR-326/hnRNPA2B1 axis, suggesting that PCAT6 functions as an oncogene and may be a useful biomarker for the future diagnosis and treatment of liver cancer.
Keywords: liver cancer, lncRNA PCAT6, miRNA-326, hnRNPA2B1
Introduction
Liver cancer is one of the most common types of cancer and is a serious threat to human health, with 854,000 new cases of liver cancer and 810,000 associated deaths globally in 2015, contributing to 20,578,000 disability-adjusted life-years (1). After surgery, the incidence of tumor recurrence and metastasis was high from 1990–2012 globally, at 33–100% (2) and the prognosis is poor (3). In recent years, surgical techniques have developed with the improvement of diagnostic methods. However, the clinical outcome of patients with unresectable disease is suboptimal with a median survival of only 12 months and no patient survived beyond 5 years from 1986–2003 in the USA (4,5). Therefore, it is important to identify novel therapeutic targets and improve the diagnosis and treatment of patients with liver cancer.
Long non-coding RNA (lncRNA) is an RNA molecule with >200 nucleotides and lacking protein-coding potential. Research has implicated lncRNAs may function as oncogene or tumor suppressor genes by regulating the transcription of protein-coding genes or altering chromatin structure (6,7). Moreover, lncRNAs have been recognized as diagnostic molecular biomarkers or therapeutic targets of cancer (8,9). For example, lncRNA H19, SNHG11 and PTCSC3 are abnormally expressed in tumor tissues and cell lines, and can serve as biomarkers for the diagnosis and prognosis of numerous solid tumors, including breast (10), colorectal (11) and gastric cancer (12). lncRNAs are involved in the development and progression of malignant tumors by regulating cancer cell proliferation, migration, invasion, apoptosis, angiogenesis and immune escape (13). Previously, prostate cancer-associated transcript 6 (PCAT6) has been identified as an lncRNA involved in the regulation of lung cancer progression (14). Subsequently, several studies have highlighted the abnormal expression of lncRNA PCAT6 in various malignant tumors, including gastrointestinal stromal tumor, colon cancer and osteosarcoma, suggesting that PCAT6 may play an essential role in tumor progression (15–17). Although the oncogenic roles of PCAT6 have been demonstrated in several tumors, little is known about the functionality and mechanism of PCAT6 in liver cancer. Moreover, evidence has shown that the exerted actions of micro (mi)RNAs depend on the modulation of upstream molecules, including lncRNAs, and downstream mRNAs (18). Evidence has suggested that microRNA (miRNA/miR) expression is abnormal in different types of tumor tissues and it is well-known that miRNAs play notable roles in tumor development and cancer therapy (19). To date, low expression levels of miR-326 have been observed in HCC tissues and cell lines, and miR-326 has been found to exert a regulatory effect on the proliferation, migration and invasion of HCC cells (20). Nevertheless, to the best of the authors knowledge, there is currently no evidence regarding the interaction between PCAT6 and miR-326 in liver cancer.
In the present study, based on the Gene Expression Profiling Interactive Analysis (GEPIA) database, the expression of lncRNA PCAT6 between liver cancer tissues and adjacent noncancerous liver tissues was analyzed. Furthermore, the functional role and mechanism of PCAT6 was explored by performing a series of experiments in vitro and in vivo. Taken together, therapeutic interventions targeting PCAT6 could be developed as a new strategy for the treatment of liver cancer.
Materials and methods
Patients and specimens
All tissue specimens (ANT, adjacent normal tissues; HCCT, liver cancer tissues) were collected from 117 patients with liver cancer, which were histologically confirmed by an experienced pathologist (GS) at The Department of Herpetological Surgery, Zhejiang Cancer Hospital (Hangzhou, China) between January 2015 and January 2017. The age range of 117 patients was 45–80 years (mean age, 58 years old), 68 were females and 49 were males. The inclusion criteria were as follows: i) ≥18 Years old; ii) histologically diagnosed with liver cancer and were amenable to surgery; iii) had never undergone any type of anti-cancer therapy, such as chemotherapy and radiotherapy, before surgery; iv) clinical data were complete. The exclusion criteria were as follows: Patients who also had other i) malignant tumors; ii) severe cardiovascular and cerebrovascular diseases and iii) hematological diseases. Tissue specimens were immediately kept in RNA Keeper Tissue Stabilizer (Vazyme Biotech Co., Ltd.) after surgical resection and then stored at −80°C. The study protocol was reviewed and approved by The Ethics Board of Zhejiang Cancer Hospital (Hangzhou, China). The study was conducted as per the principles stated in the Declaration of Helsinki and local ethical guidance documents (21). Written informed consent was obtained from all patients from whom specimens were collected.
GEPIA database
GEPIA (http://gepia.cancer-pku.cn/) is a database that provides key interactive and customizable functions, including differential expression analysis, and patient survival analysis (22). Following submission of an analysis request, GEPIA can provide the visual image results for users. The gene symbol PCAT6 and hnRNPA2B1 were submitted on GEPIA for differential expression analysis between data from liver cancer tissues and adjacent normal tissues, and provided results consisting of box plots and survival analysis. A total of 364 cases of liver cancer patients were divided into groups according to the median expression of PCAT6 or hnRNPA2B1: A high PCAT6 or hnRNPA2B1 expression group and low PCAT6 expression group. The median of relative PCAT6 expression level was 1.5, so the number of patients with high PCAT6 expression were 182 (>1.5) and the number of patients with low PCAT6 expression were 182 (≤1.5). The median of relative hnRNPA2B1 expression level was 8.2, so the number of patients with high hnRNPA2B1 expression were 182 (>8.2) and the number of patients with low hnRNPA2B1 expression were 182 (≤8.2).
Cell culture and transfection
Human liver cancer cell lines MHCC97H, HepG2 and Huh7 and the normal liver epithelial cells THLE-3 and 293T cells were obtained from The American Type Culture Collection and cultured according to their recommendations. The liver cancer cell lines were authenticated by short tandem repeat profiling.
PCAT6 short hairpin (sh)RNA and shRNA scrambled control (sh-NC), pcDNA3.1-PCAT6 and empty vector (pcDNA3.1-NC), miR-326 mimic and a scrambled negative control (NC mimic), miR-326 inhibitor (miR-Inh) and a scrambled negative control (miR-NC), hnRNPA2B1 shRNA and scrambled control (sh-NC) were purchased from GenePharm Co., Ltd. Both HepG2 and Huh7 cells were seeded into seeded in six-well plates (2×105 cells/well) and then transfected with an oligonucleotide (50 nM miRNA mimic/inhibitor) or plasmid (2 µg) using Lipofectamine® 3000 reagent (Thermo Fisher Scientific, Inc.) at 37°C for 24 h. In vivo experiments, the lentiviral vectors carrying shRNA for PCAT6 (sh-PCAT6) or negative control (sh-NC) were synthesized by RiboBio Co., Ltd.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from liver cancer tissues, adjacent non-cancerous tissues, liver cancer cell lines and normal liver epithelial cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) as described in a previous study (23). cDNA was synthesized from 1 µg of total RNA using the PrimeScript™ 1st Strand cDNA synthesis kit (Takara Bio, Inc.) according to the manufacturers instructions. qPCR was performed in an ABI 7500 instrument (Thermo Fisher Scientific, Inc.) using SYBR®-Green Real-Time PCR Master Mix (Takara Bio, Inc.) following the manufacturers protocols. The PCR reaction conditions were initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. The following primer sequences were used for qPCR: PCAT6, Forward: 5′-CAGGAACCCCCTCCTTACTC-3′ and reverse: 5′-CTAGGGATGTGTCCGAAGGA-3′; miR-326, forward: 5′-GGCGCCCAGAUAAUGCG-3′, reverse: 5′-CGTGCAGGGTCCGAGGTC-3′; hnRNPA2B1, forward: 5′-GCTGTAGCAAGAGAGGAATCTGGA-3′ and reverse: 5′-GCTTCTTCACAGTTACATGAGCCC-3′; GAPDH, forward: 5′-AGCCACATCGCTCAGACAC-3′ and reverse: 5′-GCCCAATACGACCAAATCC-3′; U6, forward: 5′-CTCGCTTCGGCAGCACA-3′ and reverse: 5′-AACGCTTCACGAATTTGCGT-3′. The expression levels of PCAT6, hnRNPA2B1 and miR-326 were calculated using the 2−ΔΔCq method (24) and normalized to GAPDH and U6 as appropriate.
Cell counting kit-8 (CCK-8) assay
After 24-h post-transfection, both HepG2 and Huh7 cells (2×103 cells/well) were seeded in 96-well plates and cultured in DMEM medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). When the density of the cells reached 70%, cells were transfected with PCAT6 shRNA and sh-NC, miR-326 mimic and NC mimic, miR-326 inhibitor and inhibitor NC, or hnRNPA2B1 shRNA and vector. After 48 h of transfection, 10 µl CCK-8 solution (Beyotime Institute of Biotechnology) was added to each well. After a 2-h incubation at 37°C, the absorbance of each well was assessed at 450 nm using a microplate reader (Thermo Fisher Scientific, Inc.).
Colony formation assay
Both HepG2 and Huh7 cells were transfected with sh-hnRNPA2B1 and sh-NC, sh-PCAT6 and sh-NC, miR-326 inhibitor and inhibitor NC, pcDNA3.1-PCAT6 and (pcDNA3.1-NC), and cultured under standard conditions. After that, the cells were diluted and seeded into six-well plates at the density of 3,000 cells per well for 14 days. The cells were then stained with 0.5% crystal violet (Beyotime Institute of Biotechnology) at 37°C for 45 min. The colony number was observed with an inverted microscope (magnification, ×10; Thermo Fisher Scientific, Inc.) and counted using ImageJ software (version 4.0; National Institutes of Health).
Western blotting
To extract total protein, both HepG2 and Huh7 cells were lysed with RIPA Lysis Buffer (Beyotime Institute of Biotechnology) as described previously (25). The lysate was centrifuged at 1,200 × g at 4°C for 10 min. The protein amount in the supernatant was quantified using a BCA assay. Subsequently, protein specimens (40 µg/lane) were added onto SDS-PAGE, and electrophoresis at 60 V. Proteins were then transferred to a polyvinylidene difluoride membrane (EMD Millipore). After blocking with 5% (w/v) non-fat dry milk at 37°C for 1 h, the membranes were incubated with primary antibodies against β-actin (1:2,000 dilution; cat. no. ab8226) and hnRNPA2B1 (1:1,000 dilution; cat. no. ab31645) at 4°C overnight. All antibodies were purchased from Abcam. Next, HRP-conjugated secondary antibodies (1:5,000 dilution; cat. no. HRP-60008, ProteinTech Group, Inc.) were added and incubated for 1 h at room temperature. The protein bands were detected using chemiluminescence (Pierce; Thermo Fisher Scientific, Inc.) and visualized on a Tanon 5200 Imaging system (Tanon Science & Technology Co., Ltd.).
Transwell chamber assay
A Transwell chamber (8-µm pore, 24-well plate; Corning, Inc.) with insert membranes coated with diluted Matrigel at 37°C for 1 h was used. HepG2 or Huh7 cells at 24-h post-transfection in serum-free medium (1×105 cells) were added to the upper chamber and cultured at 37°C for 24 h. The bottom chamber contained a medium with 10% FBS. After incubation at 37°C for 24 h, the insert membranes were cut and stained with crystal violet at 37°C for 30 min (Beijing Solarbio Science & Technology Co., Ltd.) and images were captured using an inverted microscope (magnification, ×100). The number of invading cells were counted in three wells per group.
Bioinformatics
Starbase version 2.0 (http://starbase.sysu.edu.cn/index.php) (26) were browsed to identify the miRNAs that bind lncRNA PCAT6 and hnRNPA2B1.
Dual-luciferase reporter gene assay
The wild-type (WT) sequence of PCAT6 or the hnRNPA2B1 3 untranslated region (UTR) containing binding sites of miR-326 was cloned into pmirGLO (Promega Corporation) to form the corresponding luciferase reporter vectors (WT-PCAT6 and hnRNPA2B1 3UTR-WT). The mutant (MUT)-PCAT6 and hnRNPA2B1 3UTR-MUT were obtained by mutating the seed sites. 293T cells were seeded into 24-well plates at 1×105 cells/cm2. When the cell density reached 70–80%, reporter gene plasmids and miR-326 or control vector (GenePharm Co., Ltd.) were co-transfected into 293T cells using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) at 37°C for 48 h. The relative luciferase activities were detected using the Dual-Luciferase Reporter assay kit (Promega Corporation) according to the manufacturers protocol. Luciferase activity was measured and normalized to Renilla luciferase activity. The sequences of miRNA mimic/inhibitor were as follows: miR-NC, 5′-UUCUCCGAACGUUCACGUTT-3′; miR-326 mimics, 5′-AGGAUGUCUAAAUGUUUGUUA-3′ and miR-326 inhibitor, 5′-AAGAAGUGCACCAUGUUUGUUU-3′.
Xenograft model
A total of 20 BALB/c nude mice (male; 5-weeks-old, 20–22 g) were obtained from the Hunan SJA Laboratory Animal Co., Ltd. All mice had access to food and water ad libitum, and temperature in the cages was maintained at 24±1°C with a 12/12 h light/dark cycle and 40–50% humidity. Transfected HepG2 cells (1×107) were resuspended in 100 µl PBS and subcutaneously injected into the flanks of male BALB/c mice (n=10) as previously described (27). Tumor volume was detected every 4 days and calculated as: Length (mm) × width2 (mm2)/2. After tumor growth for 28 days, mice (weight, 21–24 g) were euthanized via cervical dislocation. Tumor samples were weighed and harvested to examine the expression of miR-326 and hnRNPA2B1. The animal experiments were conducted under the approval of The Animal Care and Use Committee of Zhejiang Cancer Hospital (approval no. IRB-2019-5).
Statistical analysis
All experiments were repeated at least three times. Data are presented as the mean ± standard deviation (unless otherwise shown). The median expression of PCAT6 and hnRNPA2B1 in all human liver cancer samples were considered as the cut-off to classify groups into PCAT6- and hnRNPA2B1-high and PCAT6-and hnRNPA2B1-low expression. Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software, Inc.), and differences between groups were analyzed using one-way ANOVA and Tukeys post hoc tests. A paired or unpaired two-tailed Students t-test was used to compare differences between two groups of paired tissues or unpaired cells, respectively. The correlation relationships among PCAT6, miR-326 and hnRNPA2B1 were analyzed using the Pearsons correlation coefficient. Survival analysis was performed using log-rank tests and presented on Kaplan-Meier plots. Survival curves with cross-over were compared using the Renyi test. P<0.05 was considered to indicate a statistically significant difference.
Results
PCAT6 is upregulated and associated with the progression of liver cancer
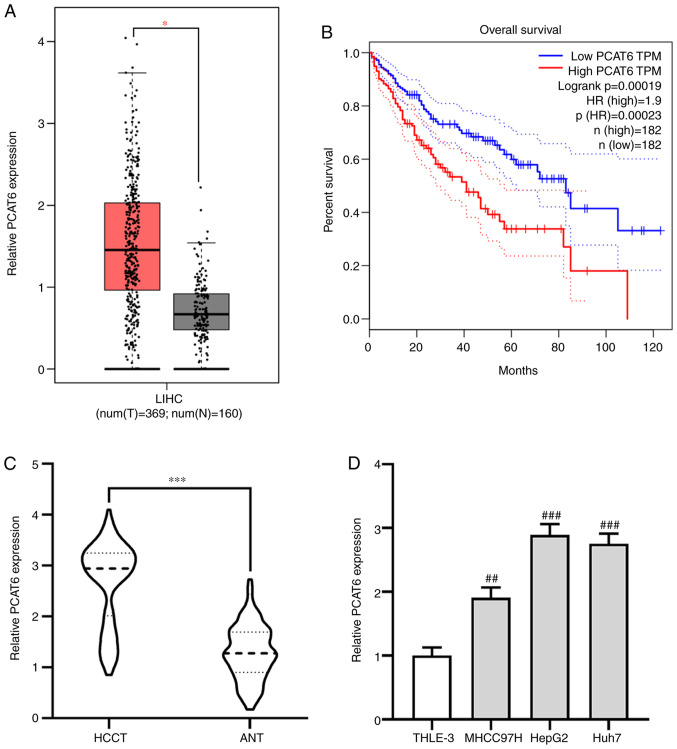
The effect of PCAT6 on the progression of liver cancer was investigated using the GEPIA database. It was found that the expression of PCAT6 was significantly elevated in liver cancer tissues (HCCT) compared with adjacent normal tissues (ANT; P<0.05; Fig. 1A). Moreover, patients with liver cancer who had a high level of PCAT6 exhibited a shorter overall survival (P<0.001; Fig. 1B). The expression pattern of PCAT6 in 117 pairs of HCCT and matching ANT was further investigated using RT-qPCR. It was found that PCAT6 expression was upregulated in HCCT compared with the ANT (P<0.001; Fig. 1C).Additionally, PCAT6 expressions in liver cancer cell lines MHCC97H, HepG2 and Huh7 were significantly enhanced compared with expression in THLE-3 normal liver epithelial cells, especially HepG2 and Huh7 cells (MHCC97H P<0.01; HepG2 and Huh7 P<0.001; Fig. 1D). Taken together, these data suggested that PCAT6 may act as an oncogene in the progression of liver cancer.
Figure 1.
PCAT6 is upregulated in liver cancer tissues and associated with clinicopathological characteristics of patients with liver cancer. (A) Expression of PCAT6 in liver cancer tissues (n=369) and normal tissues (n=160) from the GEPIA database. (B) Association between PCAT6 expression and overall survival rate of patients with liver cancer was analyzed using GEPIA database. (C) RT-qPCR was performed to examine the expression of PCAT6 in HCCT (n=117) and matched adjacent normal tissues. (D) RT-qPCR was used to detect the expression of PCAT6 in liver cancer cell lines (MHCC97H, HepG2 and Huh7) and normal liver epithelial cells THLE-3. *P<0.05 and ***P<0.001 compared with the normal tissues; ##P<0.01 and ###P<0.001 compared with the THLE-3 cell line. PCAT6, prostate cancer-associated transcript 6; GEPIA, Gene Expression Profiling Interactive Analysis; RT-qPCR, reverse transcription-quantitative PCR; HCCT, liver cancer tissue; ANT, adjacent normal tissues; LIHC, liver hepatocellular carcinoma; TPM, transcripts per million; HR, hazard ratio.
Knockdown of PCAT6 inhibits liver cancer cell proliferation and invasion
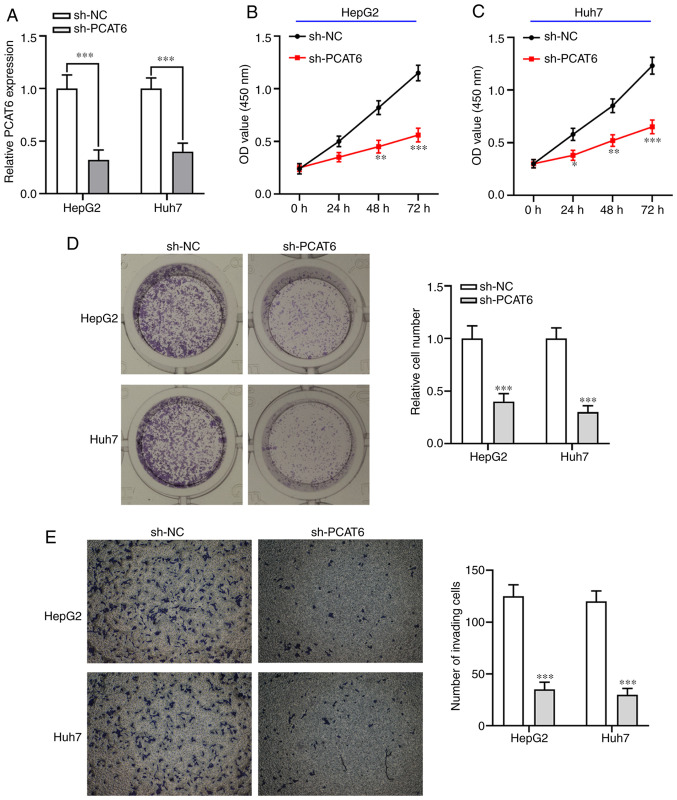
To investigate the biological functions of PCAT6 in liver cancer cells in vitro, sh-PCAT6 was transfected into both HepG2 and Huh7 cells to decrease its expression (P<0.001; Fig. 2A). The CCK-8 assay showed that knockdown of PCAT6 significantly inhibited liver cancer cell proliferation ability compared with the sh-NC group (P<0.05; Fig. 2B and C). Colony formation assays showed that PCAT6-knockdown markedly reduced the number of liver cancer cell colonies compared with the sh-NC group (P<0.001; Fig. 2D). Furthermore, inhibition of PCAT6 weakened the invasion capacity of both HepG2 and Huh7 cells (P<0.001; Fig. 2E). These data indicated that knockdown of PCAT6 exhibited an antitumor function in liver cancer by suppressing the proliferation and invasion abilities of both HepG2 and Huh7 cells.
Figure 2.
Knockdown of PCAT6 inhibits cell proliferation and invasion in liver cancer in vitro. (A) Both HepG2 and Huh7 cells were transfected with sh-PCAT6 or sh-NC and reverse transcription-quantitative PCR was used to detect the expression of PCAT6 in both cells. (B and C) Cell Counting Kit-8 assay was used to examine the cell proliferation in both HepG2 and Huh7 cell transfected with sh-PCAT6. (D) After transfection, colony formation of HepG2 and Huh7 was determined. (E) Transwell assay was applied to detect the invasive activity of liver cancer cells after transfection with sh-PCAT6 and sh-NC. *P<0.05, **P<0.01 and ***P<0.001 compared with sh-NC. PCAT6, prostate cancer-associated transcript 6; sh-, short hairpin-; NC, negative control.
miR-326 is a target gene of PCAT6
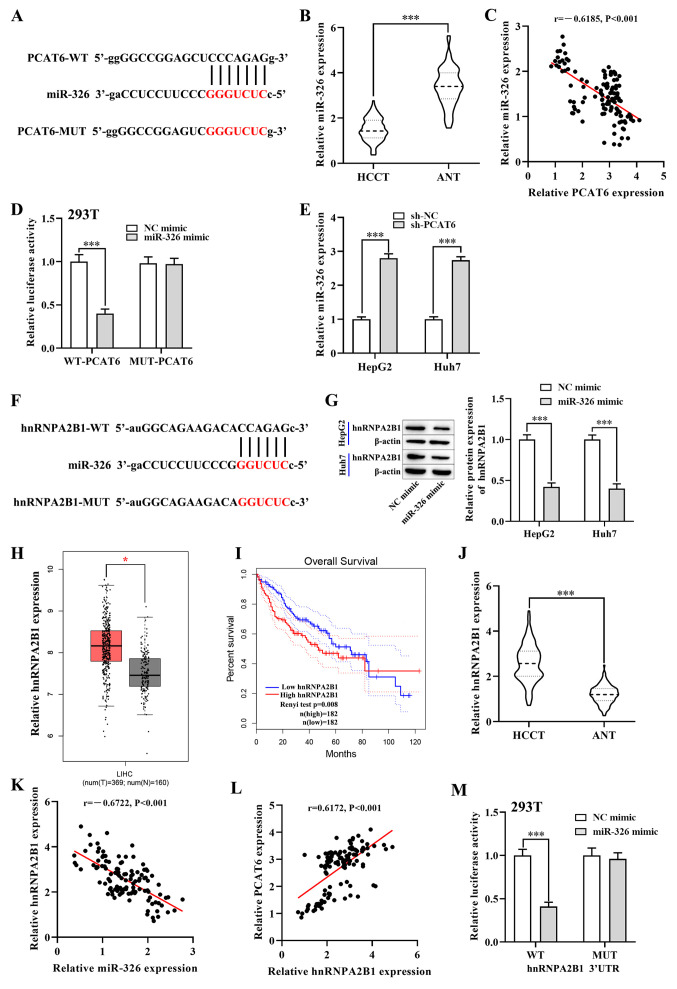
To further probe the underlying mechanism of PCAT6 function in liver cancer, the bioinformatics database, Starbase, was used to predict potential miRNAs that may be downstream target genes of PCAT6. Bioinformatics analysis showed that miR-326 could be a candidate target gene of PCAT6, as it has a potential binding site for PCAT6 (Fig. 3A). Notably, RT-qPCR revealed that the expression of miR-326 was lower in the HCCT group compared with that in the ANT group (P<0.001; Fig. 3B). A negative correlation was also found between miR-326 and PCAT6 in ANT group (P<0.001; Fig. 3C). Subsequently, the luciferase reporter assay showed that overexpression of miR-326 significantly reduced the luciferase activity of 293T cells transfected with the WT-PCAT6 vector (P<0.001; Fig. 3D), while the luciferase activity of the MUT-PCAT6 was not changed. Moreover, RT-qPCR showed that knockdown of PCAT6 markedly enhanced the expression of miR-326 in both HepG2 and Huh7 cells (P<0.001; Fig. 3E). Collectively, these results indicated that miR-326 was a target gene of PCAT6.
Figure 3.
PCAT6 promotes hnRNPA2B1 expression in liver cancer cells by targeting miR-326. (A) Starbase predicted binding sites between miR-326 and PCAT6. (B) RT-qPCR detected the expression of miR-326 in HCCT (n=117) and matched ANT. (C) Pearsons correlation analysis between PCAT6 and miR-326 in liver cancer tissues. (D) Luciferase reporter assays were performed in 293T cells to detect the binding affinity of miR-326 to the PCAT6 3UTR-WT or MUT. (E) Expression of miR-326 in both HepG2 and Huh7 cells after transfection with sh-PCAT6 or sh-NC examined using RT-qPCR. (F) Starbase database binding site predictions between miR-326 and hnRNPA2B1. (G) Protein levels of hnRNPA2B1 in both HepG2 and Huh7 cells after transfection with miR-326 mimic and NC mimic were examined using western blotting. (H) Expression of hnRNPA2B1 in liver cancer tissues (n=369) and normal tissues (n=160) from the GEPIA database. (I) Association between hnRNPA2B1 expression and overall survival rate of patients with liver cancer was analyzed using GEPIA database. (J) RT-qPCR detected the mRNA levels of hnRNPA2B1 in HCCT (n=117) and matched ANT. (K and L) Correlation between hnRNPA2B1 and PCAT6 or miR-326 in liver cancer tissues. (M) Dual-luciferase reporter assay verified the negative regulatory association between miR-326 and hnRNPA2B1 in 293T cells. *P<0.05 and ***P<0.001 compared with respective control. PCAT6, prostate cancer-associated transcript 6; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; GEPIA, Gene Expression Profiling Interactive Analysis; RT-qPCR, reverse transcription-quantitative PCR; HCCT, liver cancer tissue; ANT, adjacent normal tissue; miR-, microRNA; WT, wild-type; MUT, mutant; NC, negative control; HR, hazard ratio; LIHC; UTR, untranslated region.
Inhibition of miR-326 abrogates the inhibitory effect of PCAT6-silencing on liver cancer cell proliferation and invasion
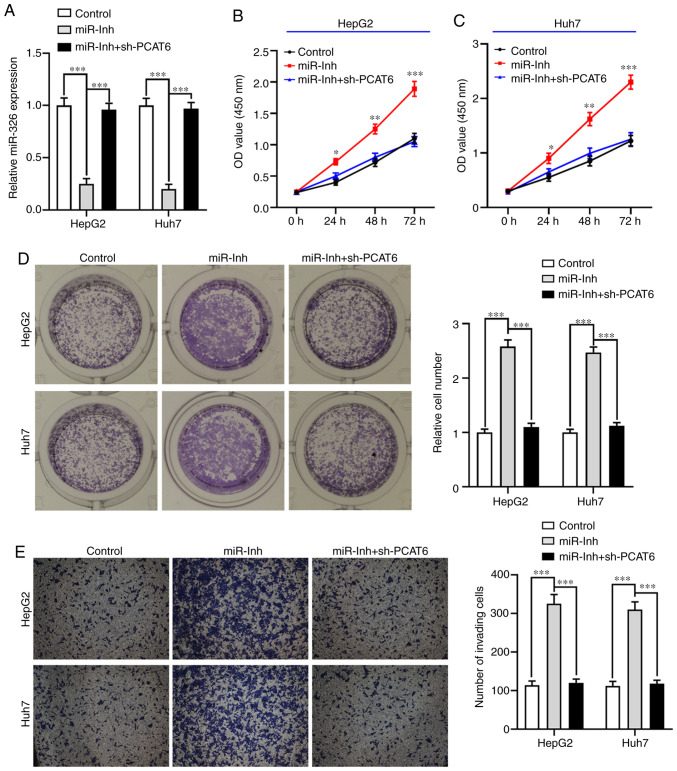
To further explore whether PCAT6 modulates the malignant phenotype of liver cancer cells by targeting miR-326, HepG2 and Huh7 cells were transfected with miR-Inh or miR-Inh + sh-PCAT6 (P<0.001; Fig. 4A). The CCK-8 assay showed that inhibition of miR-326 significantly promoted the proliferation ability of both HepG2 and Huh7 cells compared with the control group (P<0.05; Fig. 4B and C). On the other hand, knockdown of PCAT6 reversed this effect. Similarly, the clone formation of both HepG2 and Huh7 cells when transfected with miR-Inh was markedly increased compared with the control group (P<0.001; Fig. 4D), while the number of liver cancer cell colonies were reduced by knocking down both miR-326 and PCAT6. Moreover, an elevated invasive ability of both HepG2 and Huh7 cells was observed when miR-326 was suppressed, which was abrogated by inhibition of PCAT6 (P<0.001; Fig. 4E). These results demonstrated that PCAT6-silencing inhibited cell proliferation and invasion by upregulating miR-326 in liver cancer.
Figure 4.
Inhibition of miR-326 reverses the inhibitory effect of PCAT6-knockdown on liver cancer cell proliferation and invasion ability of both HepG2 and Huh7 cells. (A) Reverse transcription-quantitative PCR was used to detect the expression of miR-326 in both HepG2 and Huh7 cells. (B and C) Cell Counting Kit-8 assay was used to measure the cell proliferation in both HepG2 and Huh7 cells after transfection with miR-Inh and sh-PCAT6. (D) After transfection, HepG2 and Huh7 cell colony formation abilities were determined. (E) Transwell assays were applied to detect the invasive activity of HepG2 and Huh7 cells after transfection with miR-Inh and sh-PCAT6. *P<0.05, **P<0.01 and ***P<0.001 compared with control. miR-, microRNA; PCAT6, prostate cancer-associated transcript 6; miR-Inh, miR-326 inhibitor; sh-, short hairpin.
hnRNPA2B1 is a target of miR-326 in liver cancer cells
Next, the target of miR-326 was investigated using Starbase, which predicted that hnRNPA2B1 was a target gene (Fig. 3F). Considering the critical roles of hnRNPA2B1 in a number of malignant tumors (28), hnRNPA2B1 was selected as the target of miR-326. Notably, the GEPIA database showed that the mRNA level of hnRNPA2B1 was upregulated in liver cancer tissues compared with normal tissues (P<0.05; Fig. 3H). Moreover, the elevated expression of hnRNPA2B1 in patients with liver cancer resulted in a shorter overall survival rate (P<0.01; Fig. 3I). Likewise, the RT-qPCR analysis showed that the mRNA expression of hnRNPA2B1 in the HCCT group was higher compared with that in the ANT group (P<0.001; Fig. 3J). In addition, Pearsons correlation analysis revealed that the mRNA expression of hnRNPA2B1 was negatively correlated with miR-326 expression in HCC tumor tissues (P<0.001; Fig. 3K), while a positive correlation between hnRNPA2B1 and PCAT6 in liver cancer tissues was observed (P<0.001; Fig. 3L). To further investigate this correlation, the luciferase reporter assay showed that miR-326 mimics significantly reduced the luciferase activity of WT-hnRNPA2B1 3UTR, while this inhibition was blocked by mutation of the potential binding domains (P<0.001; Fig. 3M). Western blotting showed that the protein level of hnRNPA2B1 was significantly downregulated in both HepG2 and Huh7 cells after transfection with the miR-326 mimic (P<0.001; Figs. 3G and S1A). Overall, these results suggested that miR-326 targeted and inhibited the expression of hnRNPA2B1.
PCAT6 promotes the proliferation and invasion of liver cancer cells via the miR-326/hnRNPA2B1 axis
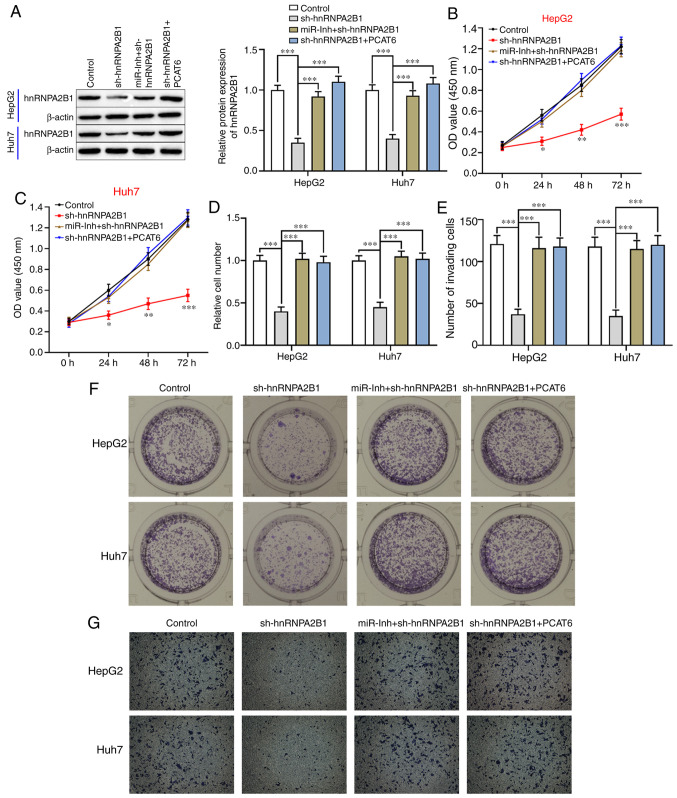
Whether PCAT6 exhibited an antitumor effect on liver cancer cells via regulation of the miR-326/hnRNPA2B1 axis was further investigated. As shown in Figs. 5A and S1B-E, the protein levels of hnRNPA2B1 were significantly reduced in both HepG2 and Huh7 cells after transfection with sh-hnRNPA2B1 (P<0.001), whereas its expression was upregulated by inhibition of miR-326 or upregulation of PCAT6 in hnRNPA2B1-silenced liver cancer cells. Silencing of hnRNPA2B1 significantly decreased the proliferation (P<0.05; Fig. 5B and C) and clone formation abilities compared with the control in both HepG2 and Huh7 cells (P<0.001; Fig. 5D and E). However, the inhibitory effect of hnRNPA2B1-silencing on liver cancer cell biological behavior was reversed by the combined downregulation of miR-326 and hnRNPA2B1 or PCAT6-overexpression and hnRNPA2B1-silencing (P<0.001, Fig. 5B-E). Moreover, the invasion capacity of HepG2 and Huh7 cells was inhibited by knockdown of hnRNPA2B1 (P<0.001; Fig. 5F and G), an effect that could be attenuated through the inhibition of miR-326 or upregulation of PCAT6. Taken together, PCAT6-inhibition contributed to decreased cell proliferation and invasion of liver cancer cells via downregulation of hnRNPA2B1 and upregulation of miR-326.
Figure 5.
PCAT6 contributes to liver cancer cell proliferation and invasion by regulating the miR-326/hnRNPA2B1 axis. (A) Western blotting was used to detect the protein level of hnRNPA2B1 in both liver cancer cell after transfection with hnRNPA2B1 shRNA (sh-hnRNPA2B1), combined hnRNPA2B1 shRNA and miR-326 inhibitor (sh-hnRNPA2B1+miR-Inh), combined hnRNPA2B1 shRNA and pcDNA3.1-PCAT6 (sh-hnRNPA2B1+PCAT6). (B and C) After transfection, the proliferation ability of both HepG2 and Huh7 cells were determined by using a Cell Counting Kit-8 assay and (D and E) colony formation assay. (F and G) Cell invasion was detected using a Transwell assay. *P<0.05, **P<0.01 and ***P<0.001. PCAT6, prostate cancer-associated transcript 6; miR-, microRNA; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; sh-, short hairpin-; miR-Inh, miR-326 inhibitor.
Knockdown of PCAT6 suppresses tumor growth of liver cancer in vivo
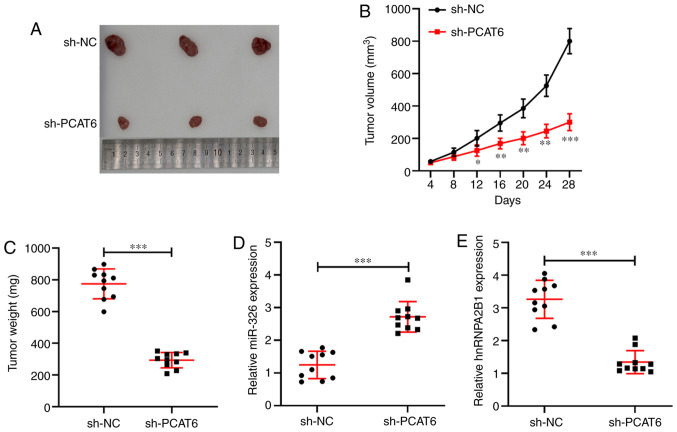
Based on the antitumor effect of PCAT6-inhibition by regulating the miR-326/hnRNPA2B1 axis in vitro, the role of PCAT6 in vivo was further examined using a subcutaneous xenograft model. As shown in Fig. 6A and C, knockdown of PCAT6 significantly decreased tumor growth and tumor weight (P<0.001) compared with the sh-NC group at 28 days. Tumor volume in the sh-PCAT6 group was significantly lower compared with in the sh-NC group (P<0.05, Fig. 6B). Moreover, RT-qPCR was performed to detect the expression of miR-326 and hnRNPA2B1 in tumor tissues derived from the xenograft mice model. PCAT6-silencing was found to enhance the expression of miR-326 while it reduced hnRNPA2B1 expression compared with the sh-NC group (P<0.001, Fig. 6D and E). Therefore, knockdown of PCAT6 inhibited tumor growth by upregulating the inhibitory effect of miR-326 on hnRNPA2B1 expression in liver cancer.
Figure 6.
Inhibition of PCAT6 reduces tumor growth of liver cancer in vivo. (A) Tumor images from the two groups of nude mice. (B) Tumor volume was measured every four days. (C) Tumor weight was detected in each group. (D and E) Reverse transcription-quantitative PCR was performed to examine the expression of miR-326 and hnRNPA2B1 in tumor tissues derived from xenograft nude mice. *P<0.05, **P<0.01 and ***P<0.001 compared with sh-NC. PCAT6, prostate cancer-associated transcript 6; miR-, microRNA; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; NC, negative control; sh-, short hairpin.
Discussion
Despite advances and progress in the field of liver cancer research, liver cancer remains a worrisome burden worldwide and the number of deaths due to liver cancer in China increased by 33.80% between in 1990–2015 (29–31). In the present study, PCAT6 was upregulated in liver cancer tissues and associated with poor outcomes in patients. Moreover, loss-of-function experiments revealed that the knockdown of PCAT6 suppressed the proliferation and invasion capacities of liver cancer cells in vitro and in vivo. Mechanistically, inhibition of PCAT6 exerted an antitumor function in liver cancer by targeting miR-326 and downregulating hnRNPA2B1 expression.
Studies have confirmed that lncRNA is a useful biomarker for the early diagnosis and prognosis of numerous diseases including cancer (32,33). For example, Sun et al (34) found five differentially expressed lncRNAs that are associated with shorter survival time in The Cancer Genome Atlas. Cai et al (35) reported that a nine lncRNA signature acts as a biomarker to predict overall survival in gastric cancer. Wan et al (36) demonstrated that serum PCAT6 could be used as a diagnostic and prognostic biomarker in patients with non-small-cell lung cancer. Moreover, lncRNAs may function as an oncogene or tumor suppressor gene involved in the progression of the malignant tumor by regulating the proliferation, invasion and migration of cancer cells (6,37). For instance, Wang et al (38) demonstrated that a novel lncRNA MCM3AP antisense RNA 1 facilitates tumor growth of HCC by promoting HCC cell proliferation and reducing apoptosis. In the present study, PCAT6 was shown to be overexpressed in liver cancer tissues and cell lines, and inhibition of PCAT6 significantly decreased liver cancer cell proliferation and invasion. Of note, a recent study reported that PCAT6 serves as an oncogene to promote the development of non-small-cell lung cancer by binding to the enhancer of zeste homolog 2 (EZH2) and suppressing the expression of large tumor suppressor homolog 2 (39). Lv et al (40) showed that PCAT6-overexpression promotes cell proliferation, migration, invasion and reduces apoptosis through the activation of the Wnt/β-catenin pathway in cervical cancer. Other studies have demonstrated that knockdown of PCAT6 reverses chemoresistance of cancer cells to 5-fluorouracil and enhances radiosensitivity in breast cancer cells (41,42). The aforementioned results of the present study indicated that the oncogene PCAT6 might be an effective therapeutic target for patients with liver cancer.
Numerous studies have confirmed that lncRNA plays a major role in regulating cancer progression by targeting miRNA to regulate mRNA expression (43). For example, Dong et al (44) reported that silencing of PCAT6 restrains cell proliferation and epithelial-mesenchymal transition of gastric cancer cells by targeting miR-15a. Huang et al (16) confirmed that PCAT6 suppresses apoptosis and contributes to cell proliferation by upregulating anti-apoptotic protein apoptosis repressor with caspase recruitment domain via EZH2 in colon cancer. Zhu et al (17) reported that PCAT6 promotes tumor progression in osteosarcoma by regulating the miR-185-5p/TGFBR1/2 axis. The results of the present study showed that PCAT6 targeted miR-326 to promote tumorigenesis of liver cancer by upregulating the expression of hnRNPA2B1. Notably, a number of studies have demonstrated that miR-326 acts as a tumor suppressor in cancer progression (45,46). Lu et al (47) reported that miR-326 is a novel miRNA signature for HCC diagnosis and prognosis. Meanwhile, evidence suggests that miR-326 is involved in malignant phenotypes by targeting downstream genes or signaling pathways. For instance, overexpression of miR-326 targets LIM and SH3 protein 1 to suppress HCC cell proliferation and invasion and promote apoptosis (48). Mo et al (49) demonstrated that miR-326 inhibits HCC tumor growth by blocking the Akt/c-Myc pathway through targeting of 3-phosphoinositide-dependent protein kinase 1. Upregulation of miR-326 reduces chemoresistance in numerous solid tumors, such as HCC (50), gastric cancer (51) and lung adenocarcinoma (52). In the present study, miR-326 directly targeted hnRNPA2B1, which is reported as an oncogene in HCC (53). In the present study, knockdown of hnRNPA2B1 significantly suppressed liver cancer cell proliferation and invasion, which is also in agreement with a previous study by Wang et al (54). Furthermore, previous studies demonstrated that silencing of hnRNPA2B1 inhibits malignant capability and induces apoptosis through downregulation of lin28B and inactivation of the PI3K/Akt pathway in tumors (55,56). Notably, the data from the present study showed that inhibition of miR-326 or upregulation of PCAT6 significantly alleviated the inhibitory effect of hnRNPA2B1 deletion on the proliferation and invasion of liver cancer cells.
It was concluded that PCAT6 is upregulated in liver cancer tissues and significantly associated with poor prognosis in patients with liver cancer. Loss-of-function experiments demonstrated that knockdown of PCAT6 exhibited an anti-tumorigenic effect in liver cancer by downregulating hnRNPA2B1 expression through the targeting of miR-326. The study provided a theoretical basis for PCAT6 as a new diagnostic, prognostic and therapeutic target for patients with liver cancer.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ANT
adjacent normal tissue
- CCK-8
Cell Counting Kit-8
- GEPIA
Gene Expression Profiling Interactive Analysis
- HCCT
liver cancer tissue
- HCC
hepatocellular carcinoma
- hnRNPA2B1
heterogeneous nuclear ribonucleoprotein A2B1
- PCAT6
prostate cancer-associated transcript 6
- MUT
mutant type
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- TCGA
The Cancer Genome Atlas
- WT
wild-type
Funding Statement
This work was supported by The Medicine and Health Discipline Platform Project of Zhejiang Province (grant no. 2018RC019) and The Medicine and Health Science Project of Zhejiang Province (grant no. 2020KY483).
Funding
This work was supported by The Medicine and Health Discipline Platform Project of Zhejiang Province (grant no. 2018RC019) and The Medicine and Health Science Project of Zhejiang Province (grant no. 2020KY483).
Availability of data and materials
The datasets used and/or analyzed in the current study are available from the corresponding author on request.
Authors contributions
JL and GS designed the experiments. JL, JZ and WH performed experiments. WH, HZ and ZZ analyzed the data. JZ and WH confirmed the authenticity of all raw data. JL and JZ drafted the manuscript. GS revised manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by The Ethics Committee of Zhejiang Cancer Hospital (Hangzhou, China; approval no. IRB-2020-6). All patients have signed written informed consent. The animal study was approved by the Animal Care and Use Committee of the Zhejiang Cancer Hospital (approval no. IRB-2019-5).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Global Burden of Disease Liver Cancer Collaboration. Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, Allen C, Al-Raddadi R, Alvis-Guzman N, Amoako Y, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: Results from the global burden of disease study 2015. JAMA Oncol. 2017;3:1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mavros MN, Mayo SC, Hyder O, Pawlik TM. A systematic review: Treatment and prognosis of patients with fibrolamellar hepatocellular carcinoma. J Am Coll Surg. 2012;215:820–830. doi: 10.1016/j.jamcollsurg.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Tan N, Zhu B, Shu H, Tao YF, Wu JR, Fang M, Li CR, Chen ZQ, Ou C. Effect of lncRNA-BC200 on proliferation and migration of liver cancer cells in vitro and in vivo. Oncol Rep. 2020;43:461–470. doi: 10.3892/or.2019.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stipa F, Yoon SS, Liau KH, Fong Y, Jarnagin WR, DAngelica M, Abou-Alfa G, Blumgart LH, DeMatteo RP. Outcome of patients with fibrolamellar hepatocellular carcinoma. Cancer. 2006;106:1331–1338. doi: 10.1002/cncr.21703. [DOI] [PubMed] [Google Scholar]
- 5.Kakar S, Burgart LJ, Batts KP, Garcia J, Jain D, Ferrell LD. Clinicopathologic features and survival in fibrolamellar carcinoma: Comparison with conventional hepatocellular carcinoma with and without cirrhosis. Mod Pathol. 2005;18:1417–1423. doi: 10.1038/modpathol.3800449. [DOI] [PubMed] [Google Scholar]
- 6.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24:257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Feng W, Liu W, Kong X, Li L, He J, Wang D, Zhang M, Zhou G, Xu W, et al. Circulating lncRNA ABHD11-AS1 serves as a biomarker for early pancreatic cancer diagnosis. J Cancer. 2019;10:3746–3756. doi: 10.7150/jca.32052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8:1015. doi: 10.3390/cells8091015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong G, Wang K, Li J, Xiao S, Wei W, Liu J. Determination of serum exosomal H19 as a noninvasive biomarker for breast cancer diagnosis. Onco Targets Ther. 2020;13:2563–2571. doi: 10.2147/OTT.S243601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu W, Zhou G, Wang H, Liu Y, Chen B, Chen W, Lin C, Wu S, Gong A, Xu M. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer. 2020;146:2901–2912. doi: 10.1002/ijc.32747. [DOI] [PubMed] [Google Scholar]
- 12.Zhang G, Chi N, Lu Q, Zhu D, Zhuang Y. LncRNA PTCSC3 is a biomarker for the treatment and prognosis of gastric cancer. Cancer Biother Radiopharm. 2020;35:77–81. doi: 10.1089/cbr.2019.2991. [DOI] [PubMed] [Google Scholar]
- 13.Yao Z, Zhang Y, Xu D, Zhou X, Peng P, Pan Z, Xiao N, Yao J, Li Z. Research progress on long non-coding RNA and radiotherapy. Med Sci Monit. 2019;25:5757–5770. doi: 10.12659/MSM.915647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Knockdown of long noncoding RNA PCAT6 inhibits proliferation and invasion in lung cancer cells. Oncol Res. 2016;24:161–170. doi: 10.3727/096504016X14618564639178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai F, Zhang N, Fang W, He X, Zheng Y, Gu D. PCAT6 mediates cellular biological functions in gastrointestinal stromal tumor via upregulation of PRDX5 and activation of Wnt pathway. Mol Carcinog. 2020;59:661–669. doi: 10.1002/mc.23199. [DOI] [PubMed] [Google Scholar]
- 16.Huang W, Su G, Huang X, Zou A, Wu J, Yang Y, Zhu Y, Liang S, Li D, Ma F, Guo L. Long noncoding RNA PCAT6 inhibits colon cancer cell apoptosis by regulating anti-apoptotic protein ARC expression via EZH2. Cell Cycle. 2019;18:69–83. doi: 10.1080/15384101.2018.1558872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu C, Huang L, Xu F, Li P, Li P, Hu F. LncRNA PCAT6 promotes tumor progression in osteosarcoma via activation of TGF-β pathway by sponging miR-185-5p. Biochem Biophys Res Commun. 2020;521:463–470. doi: 10.1016/j.bbrc.2019.10.136. [DOI] [PubMed] [Google Scholar]
- 18.Tang X, Feng D, Li M, Zhou J, Li X, Zhao D, Hao B, Li D, Ding K. Transcriptomic analysis of mRNA-lncRNA-miRNA interactions in hepatocellular carcinoma. Sci Rep. 2019;9:16096. doi: 10.1038/s41598-019-52559-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gnoni A, Santini D, Scartozzi M, Russo A, Licchetta A, Palmieri V, Lupo L, Faloppi L, Palasciano G, Memeo V, et al. Hepatocellular carcinoma treatment over sorafenib: Epigenetics, microRNAs and microenvironment. Is there a light at the end of the tunnel? Expert Opin Ther Targets. 2015;19:1623–1635. doi: 10.1517/14728222.2015.1071354. [DOI] [PubMed] [Google Scholar]
- 20.He S, Yang J, Jiang S, Li Y, Han X. Circular RNA circ_0000517 regulates hepatocellular carcinoma development via miR-326/IGF1R axis. Cancer Cell Int. 2020;20:404. doi: 10.1186/s12935-020-01496-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Council for International Organizations of Medical Sciences (CIOMS), corp-author CIOMS; Geneva: 2002. International Ethical Guidelines for Biomedical Research Involving Human Subjects; p. p60. [PubMed] [Google Scholar]
- 22.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng J, Luo J, Zeng H, Guo L, Shao G. 125I suppressed the Warburg effect via regulating miR-338/PFKL axis in hepatocellular carcinoma. Biomed Pharmacother. 2019;119:109402. doi: 10.1016/j.biopha.2019.109402. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Zeng H, Zheng J, Wen S, Luo J, Shao G, Zhang Y. MicroRNA-339 inhibits human hepatocellular carcinoma proliferation and invasion via targeting ZNF689. Drug Des Devel Ther. 2019;13:435–445. doi: 10.2147/DDDT.S186352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li HW, Liu J. Circ_0009910 promotes proliferation and metastasis of hepatocellular carcinoma cells through miR-335-5p/ROCK1 axis. Eur Rev Med Pharmacol Sci. 2020;24:1725–1735. doi: 10.26355/eurrev_202002_20349. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Shi SL. The roles of hnRNP A2/B1 in RNA biology and disease. Wiley Interdiscip Rev RNA. 2021;12:e1612. doi: 10.1002/wrna.1612. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Zhang Y, Chen Y, Ding L. Incidence trend of liver cancer: An analysis of 40 years data from Qidong population-based cancer registry. Zhongguo Zhong Liu. 2014;23:621–628. [Google Scholar]
- 31.Sun Z, Chen T, Thorgeirsson SS, Zhan Q, Chen J, Park JH, Lu P, Hsia CC, Wang N, Xu L, et al. Dramatic reduction of liver cancer incidence in young adults: 28 year follow-up of etiological interventions in an endemic area of China. Carcinogenesis. 2013;34:1800–1805. doi: 10.1093/carcin/bgt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108:1927–1933. doi: 10.1111/cas.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camacho CV, Choudhari R, Gadad SS. Long noncoding RNAs and cancer, an overview. Steroids. 2018;133:93–95. doi: 10.1016/j.steroids.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Sun Y, Zhang F, Wang L, Song X, Jing J, Zhang F, Yu S, Liu H. A five lncRNA signature for prognosis prediction in hepatocellular carcinoma. Mol Med Rep. 2019;19:5237–5250. doi: 10.3892/mmr.2019.10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai C, Yang L, Tang Y, Wang H, He Y, Jiang H, Zhou K. Prediction of overall survival in gastric cancer using a nine-lncRNA. DNA Cell Biol. 2019;38:1005–1012. doi: 10.1089/dna.2019.4832. [DOI] [PubMed] [Google Scholar]
- 36.Wan L, Zhang L, Fan K, Wang JJ. Diagnostic significance of circulating long noncoding RNA PCAT6 in patients with non-small cell lung cancer. Onco Targets Ther. 2017;10:5695–5702. doi: 10.2147/OTT.S149314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu Q, Tong X, Yang W, Xu Q, Huang D, Tu K. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18:28. doi: 10.1186/s12943-019-0957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi X, Liu Z, Liu Z, Feng X, Hua F, Hu X, Wang B, Lu K, Nie F. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177–187. doi: 10.1016/j.ebiom.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lv XJ, Tang Q, Tu YQ, Yan DD, Wei QC. Long noncoding RNA PCAT6 regulates cell growth and metastasis via Wnt/β-catenin pathway and is a prognosis marker in cervical cancer. Eur Rev Med Pharmacol Sci. 2019;23:1947–1956. doi: 10.26355/eurrev_201903_17233. [DOI] [PubMed] [Google Scholar]
- 41.Wu H, Zou Q, He H, Liang Y, Lei M, Zhou Q, Fan D, Shen L. Long non-coding RNA PCAT6 targets miR-204 to modulate the chemoresistance of colorectal cancer cells to 5-fluorouracil-based treatment through HMGA2 signaling. Cancer Med. 2019;8:2484–2495. doi: 10.1002/cam4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi R, Wu P, Liu M, Chen B, Cong L. Knockdown of lncRNA PCAT6 enhances radiosensitivity in triple-negative breast cancer cells by regulating miR-185-5p/TPD52 axis. Onco Targets Ther. 2020;13:3025–3037. doi: 10.2147/OTT.S237559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin C, Yuan G, Hu Z, Zeng Y, Qiu X, Yu H, He S. Bioinformatics analysis of the interactions among lncRNA, miRNA and mRNA expression, genetic mutations and epigenetic modifications in hepatocellular carcinoma. Mol Med Rep. 2019;19:1356–1364. doi: 10.3892/mmr.2018.9728. [DOI] [PubMed] [Google Scholar]
- 44.Dong D, Lun Y, Sun B, Sun H, Wang Q, Yuan G, Quan J. Silencing of long non-coding RNA PCAT6 restrains gastric cancer cell proliferation and epithelial-mesenchymal transition by targeting microRNA-15a. Gen Physiol Biophys. 2020;39:1–12. doi: 10.4149/gpb_2019044. [DOI] [PubMed] [Google Scholar]
- 45.Zou X, Zhong J, Li J, Su Z, Chen Y, Deng W, Li Y, Lu S, Lin Y, Luo L, et al. miR-362-3p targets nemo-like kinase and functions as a tumor suppressor in renal cancer cells. Mol Med Rep. 2016;13:994–1002. doi: 10.3892/mmr.2015.4632. [DOI] [PubMed] [Google Scholar]
- 46.Wang D, Wang H, Li Y, Li Q. MiR-362-3p functions as a tumor suppressor through targeting MCM5 in cervical adenocarcinoma. Biosci Rep. 2018;38:BSR20180668. doi: 10.1042/BSR20180668. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Lu M, Kong X, Wang H, Huang G, Ye C, He Z. A novel microRNAs expression signature for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:8775–8784. doi: 10.18632/oncotarget.14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu S, Ran Y, Chen W, Zhang Y, Xu Y. MicroRNA-326 inhibits cell proliferation and invasion, activating apoptosis in hepatocellular carcinoma by directly targeting LIM and SH3 protein 1. Oncol Rep. 2017;38:1569–1578. doi: 10.3892/or.2017.5810. [DOI] [PubMed] [Google Scholar]
- 49.Mo Y, He L, Lai Z, Wan Z, Chen Q, Pan S, Li L, Li D, Huang J, Xue F, Che S. Gold nano-particles (AuNPs) carrying miR-326 targets PDK1/AKT/c-myc axis in hepatocellular carcinoma. Artif Cells Nanomed Biotechnol. 2019;47:2830–2837. doi: 10.1080/21691401.2018.1489266. [DOI] [PubMed] [Google Scholar]
- 50.Ma J, Wang T, Guo R, Yang X, Yin J, Yu J, Xiang Q, Pan X, Tang H, Lei X. Involvement of miR-133a and miR-326 in ADM resistance of HepG2 through modulating expression of ABCC1. J Drug Target. 2015;23:519–524. doi: 10.3109/1061186X.2015.1015536. [DOI] [PubMed] [Google Scholar]
- 51.Song W, Qian Y, Zhang MH, Wang H, Wen X, Yang XZ, Dai WJ. The long non-coding RNA DDX11-AS1 facilitates cell progression and oxaliplatin resistance via regulating miR-326/IRS1 axis in gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24:3049–3061. doi: 10.26355/eurrev_202003_20669. [DOI] [PubMed] [Google Scholar]
- 52.Yu W, Peng W, Sha H, Li J. Hsa_circ_0003998 promotes chemoresistance via modulation of miR-326 in lung adenocarcinoma cells. Oncol Res. 2019;27:623–628. doi: 10.3727/096504018X15420734828058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizuno H, Honda M, Shirasaki T, Yamashita T, Yamashita T, Mizukoshi E, Kaneko S. Heterogeneous nuclear ribonucleoprotein A2/B1 in association with hTERT is a potential biomarker for hepatocellular carcinoma. Liver Int. 2012;32:1146–1155. doi: 10.1111/j.1478-3231.2012.02778.x. [DOI] [PubMed] [Google Scholar]
- 54.Wang H, Liang L, Dong Q, Huan L, He J, Li B, Yang C, Jin H, Wei L, Yu C, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics. 2018;8:2814–2829. doi: 10.7150/thno.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y, Wei Q, Tang Y, Wang Y, Luo Q, Zhao H, He M, Wang H, Zeng Q, Lu W, et al. Loss of hnRNPA2B1 inhibits malignant capability and promotes apoptosis via down-regulating Lin28B expression in ovarian cancer. Cancer Lett. 2020;475:43–52. doi: 10.1016/j.canlet.2020.01.029. [DOI] [PubMed] [Google Scholar]
- 56.Shi X, Ran L, Liu Y, Zhong SH, Zhou PP, Liao MX, Fang W. Knockdown of hnRNP A2/B1 inhibits cell proliferation, invasion and cell cycle triggering apoptosis in cervical cancer via PI3K/AKT signaling pathway. Oncol Rep. 2018;39:939–950. doi: 10.3892/or.2018.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed in the current study are available from the corresponding author on request.