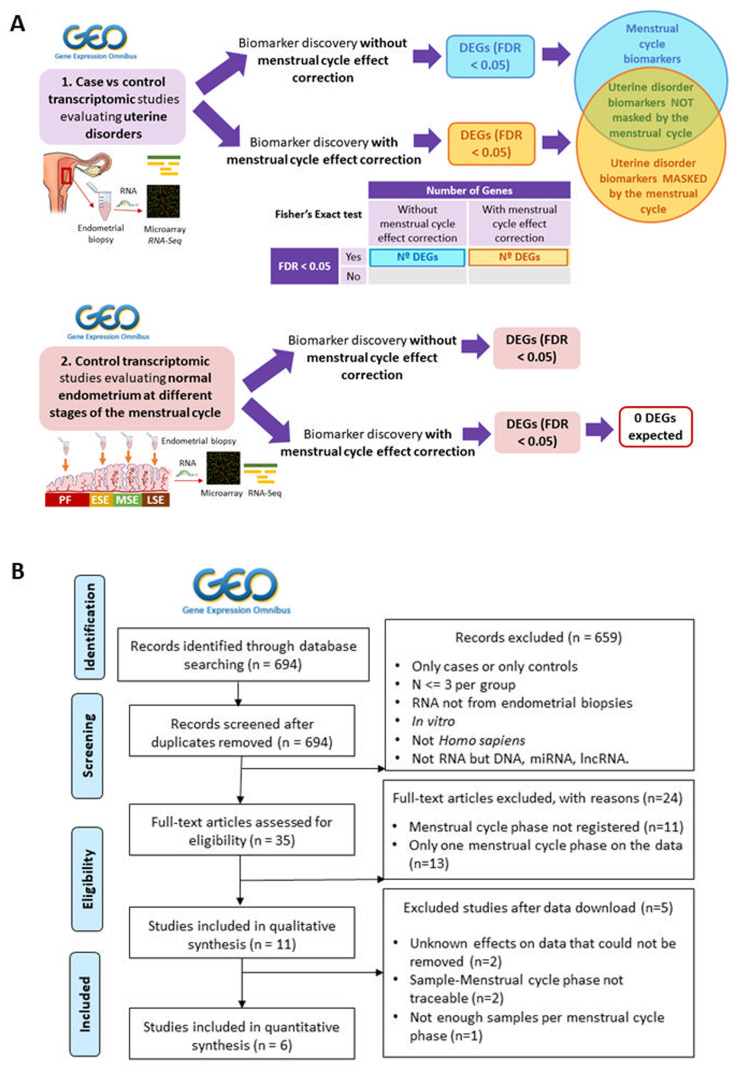
Figure 1.
(A) Study design and flowchart for transcriptomic studies evaluating uterine disorders. Systematic search and review of (i) transcriptomic case versus control studies evaluating uterine disorders and (ii) transcriptomic studies evaluating menstrual cycle progression in control women with normal endometrium were performed at the Gene Expression Omnibus (GEO) database. After filtering and selection, raw data were downloaded and pre-processed. For each included study, a differential expression analysis was applied with and without removing the menstrual cycle effect on the data using linear models. For case versus control studies (i), the proportions of differentially expressed genes (DEGs; FDR < 0.05) obtained under both approaches were compared using a Fisher’s Exact test and distinct types of endometrial biomarkers were established. Control studies evaluating menstrual cycle progression (ii) were used to evaluate the reliability of menstrual cycle effect correction; if the method properly removed the menstrual cycle effect from transcriptomic data, the differential expression analysis between endometrial phases after correcting this effect should indicate no differentially expressed genes. (B) Flowchart of the selection process of case versus control transcriptomic studies evaluating uterine disorders. The selection of suitable individual transcriptomic studies at GEO and the number of individual studies excluded and remaining after each filtering step are shown. GEO, Gene Expression Omnibus. n, number of studies. N, sample size. lncRNA, long noncoding RNA.