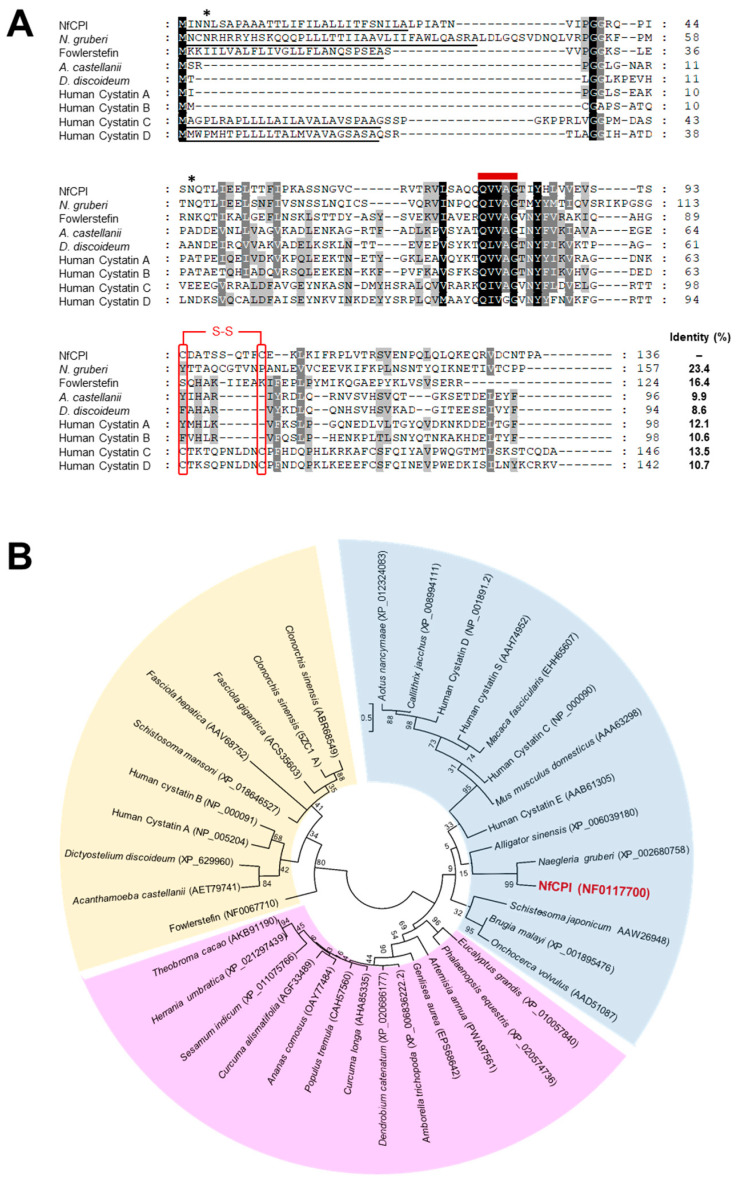
Figure 1.
Multiple sequence alignment and phylogenetic analysis. (A) Multiple sequence alignment. The deduced amino acid sequence of N. fowleri (NfCPI) was aligned with the sequences of related proteins derived from other protozoa and humans. The Gln-Val-Val-Ala-Gly (QVVAG) cystatin motif is presented as a bold red line on the sequences. The predicted N-terminal signal peptide sequences are underlined. The predicted putative N-glycosylation sites in NfCPI are marked by asterisks. A potential disulfide bridge is presented in brackets and the associated cysteine residues are boxed in red color. The shading displays the degree of identity among the sequences. Sequence identity between NfCPI and other related proteins is shown on the right side. Sequence identity between NfCPI and other related proteins is shown on the right side. Naegleria gruberi CPI (XP_002680758.1), Fowlerstefin (NF0067710), Acanthamoeba castellanii CPI (AET79741), Dictyostelium discoideum CPI (XP_629960), Human cystatin A (NP_005204), Human cystatin B (NM_000100), Human cystatin C (NP_000090), and Human cystatin D (NP_001891) are included in the alignment. (B) The phylogenetic tree was constructed by the maximum likelihood method using the MEGA6 program. Yellow, cystatin A and B clades; blue, cystatin C, D, and S clades; pink, CPIs from plants. Numbers on the branches show bootstrap proportion (1000 replicates).