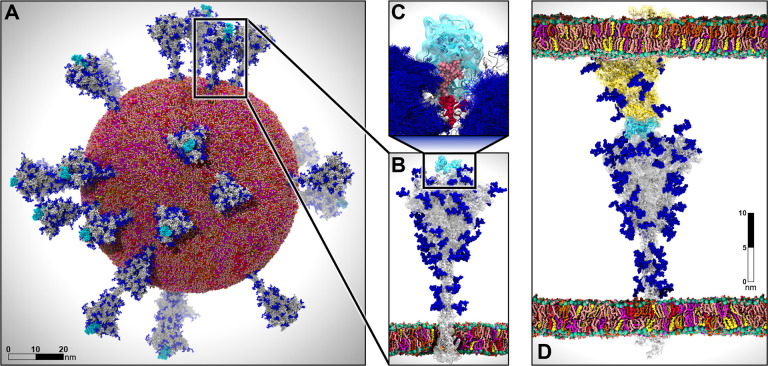
Figure 1.
Multiscale modeling of SARS-CoV-2. A) All-atom model of the SARS-CoV-2 viral envelope (305 M atoms), including 24 spike proteins (colored in gray) in both the open (16) and closed states (8). The RBDs in the “up” state are highlighted in cyan) N-/O-Glycans are shown in blue. Water molecules and ions have been omitted for clarity. B) Full-length model of the glycosylated SARS-CoV-2 spike protein (gray surface) embedded into an ERGIC-like lipid bilayer (1.7 M atoms). RBD in the “up” state is highlighted in cyan. C) The glycan shield is shown by overlaying multiple conformations for each glycan collected at subsequent timesteps along the dynamics (blue bushlike representation). Highlighted in pink and red are two N-glycans (linked to N165 and N234, respectively) responsible for the modulation of the RBD dynamics, thus priming the virus for infection. The RBD “up” is depicted with a cyan surface. D) Two-parallel-membrane system of the spike-ACE2 complex (8.5 M atoms). The spike protein, embedded into an ERGIC-like membrane, is depicted with a gray transparent surface, whereas ACE2 is shown with a yellow transparent surface and it is embedded into a lipid bilayer mimicking the composition of mammalian cell membranes. Glycans are shown in blue, whereas water has been omitted for clarity. Visualizations were created in VMD using its custom GPU-accelerated ray tracing engine (Humphrey et al., 1996; Stone et al., 2013a, 2013b, 2016a, 2016b).