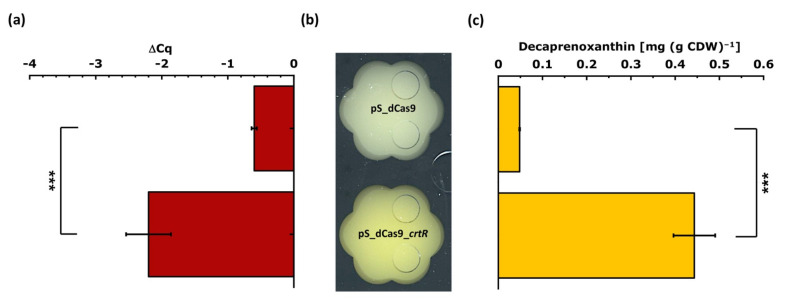
Figure 2.
Testing the CRISPRi system for identification of metabolic engineering targets relevant for carotenoid production. Strain C. glutamicum MB001(pS_dCas9_crtR) for CRISPRi-mediated repression of crtR was compared to the empty vector carrying control strain MB001(pS_dCas9) with respect to (a) crtR RNA levels, (b) color phenotypes, and (c) cellular decaprenoxanthin content. For qRT-PCR analysis, exponentially growing cells were harvested and sigA was used as a reference (a). The color phenotype (b) was judged by visual inspection after growth in the Biolector®flowerplate microcultivation system. Cells were grown in 40 g L−1 of glucose CGXII minimal medium for 28 h and induced at 0 h with 1 mM IPTG and 0.25 µg mL−1 of aTc. The cellular decaprenoxanthin content (c) is given as ß-carotene equivalents, as determined by HPLC analysis. Mean values and standard deviations of three biological replicates are given. The p-value of <0.001 (***) was calculated using Student’s t-test (two sided, unpaired).