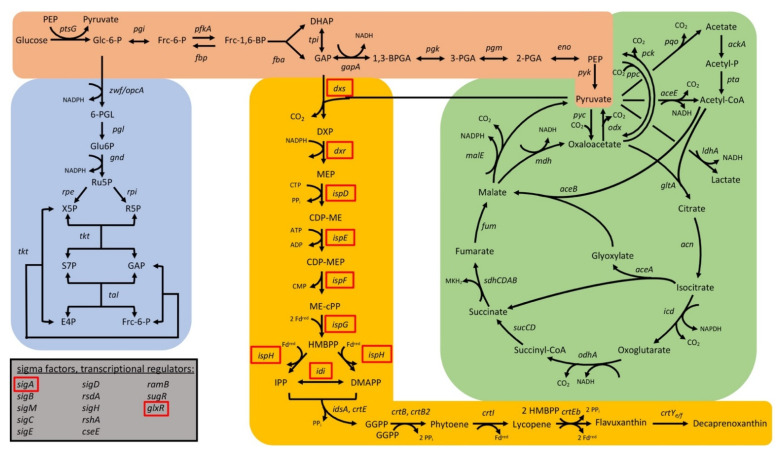
Figure 3.
Scheme of the central carbon metabolism and carotenogenesis in C. glutamicum with glycolysis (orange shading), the pentose phosphate pathway (blue shading), the TCA cycle (green shading), as well as the MEP pathway and the carotenogenesis (yellow shading). Gene names are given next to the reactions catalyzed by their gene products. The corresponding gene identifiers can be found in Table 1. Essential genes are depicted with a red box. aceA: isocitrate lyase; aceB: malate synthase; aceE: pyruvate dehydrogenase E1 component; ackA: acetate kinase; acn: aconitase; crtB: phytoene synthase; crtB2: phytoene synthase 2; crtE: geranylgeranyl-diphosphate synthase; crtEb: lycopene elongase; crtI: phytoene desaturase; crtI2: phytoene desaturase; crtYe/f: C50 carotenoid epsilon cyclase; cseE: anti-sigma factor E; dxr: 1-deoxy-D-xylulose 5-phosphate reductoisomerase; dxs: 1-deoxyxylulose-5-phosphate synthase; eno: enolase; fba: fructose-1,6-bisphosphate aldolase; fbp: fructose 1,6-bisphosphatase; fum: fumarase; gapA: glyceraldehyde-3-phosphate dehydrogenase A; gltA: citrate synthase; glxR: global transcriptional regulator; gnd: 6-phosphogluconate dehydrogenase; icd: isocitrate dehydrogenase; idi: isopentenyldiphosphate isomerase; idsA: geranylgeranyl diphosphate synthase; ispD: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; ispE: 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; ispF: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; ispG: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase; ispH: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; ldh: NAD-dependent L-lactate dehydrogenase; malE: malic enzyme; mdh: malate dehydrogenase; odhA: oxoglutarate dehydrogenase subunit A; odx: oxaloacetate decarboxylase; opcA: glucose-6-phosphate dehydrogenase; pck: phosphoenolpyruvate carboxykinase; pfkA: 6-phosphofructokinase; pgi: glucose-6-phosphate isomerase; pgk: phosphoglycerate kinase; pgl: 6-phosphogluconolactonase; pgm: phosphoglucomutase; ppc: phosphoenolpyruvate carboxylase; pqo: pyruvate quinone oxidoreductase; pta: phosphotransacetylase; ptsG: glucose-specific enzyme II BC component of PTS; pyc: pyruvate carboxylase; pyk: pyruvate kinase; ramB: transcriptional regulator of acetate metabolism A; rpe: ribulose-5-phosphate epimerase; rpi: phosphopentose isomerase; rsdA: anti-sigma factor D; rshA: anti-sigma factor H; sdhABDC: succinate dehydrogenase subunits A, B, C, and D; sigA: sigma factor A; sigB: sigma factor B; sigC: sigma factor C; sigD: sigma factor D; sigE: sigma factor E; sigH: sigma factor H; sigM: sigma factor M; sucCD: succinyl-CoA synthetase beta and alpha subunits; sugR: transcriptional regulators of sugar metabolism; tal: transaldolase; tkt: transketolase; tpi: triosephosphate isomerase; zwf: glucose-6-phosphate 1-dehydrogenase. 1,3-BPGA: 1,3-bisphosphate glycerate; 2-PGA:2-phosphate glycerate; 3-PGA: 3-phosphate glycerate; 6-PGI: 6-phosphogluconolactone; acetyl-P: acetyl-phosphate; CDP-ME: 4-diphosphocytidyl-2-methylerythritol; CDP-MEP: 4-diphosphocytidyl-2-methylerythritol 2-phosphate; DHAP: dihydroxyacetone phosphate; DMAPP: dimethylallyl diphosphate; DXP: 1-deoxy-D-xylulose 5-phosphate synthase; E4P: erythrose-4-phosphate; Frc-1,6-BP: fructose-1,6-bisphosphate; Frc-6-P: fructose-6-phosphate; GAP: glyceraldehyde 3-phosphate; GGPP: geranylgeranyl diphosphate; Glc-6-P: glucose-6-phosphate; Glu6P: 6-phosphogluconate; HMBPP: (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate; IPP: isopentenyl diphosphate; ME-cPP: 2-methylerythritol 2,4-cyclodiphosphate; MEP: 2-methylerythritol 4-phosphate; PEP: phosphoenolpyruvate; R5P: ribose-5-phosphate; Ru5P: ribulose-5-phosphate; S7P: sedoheptulose 7-phosphate;X5P: xylulose-5-phosphate.