Figure 1. Schematic representation of conformational landscape associated with steps of enzyme catalysis.
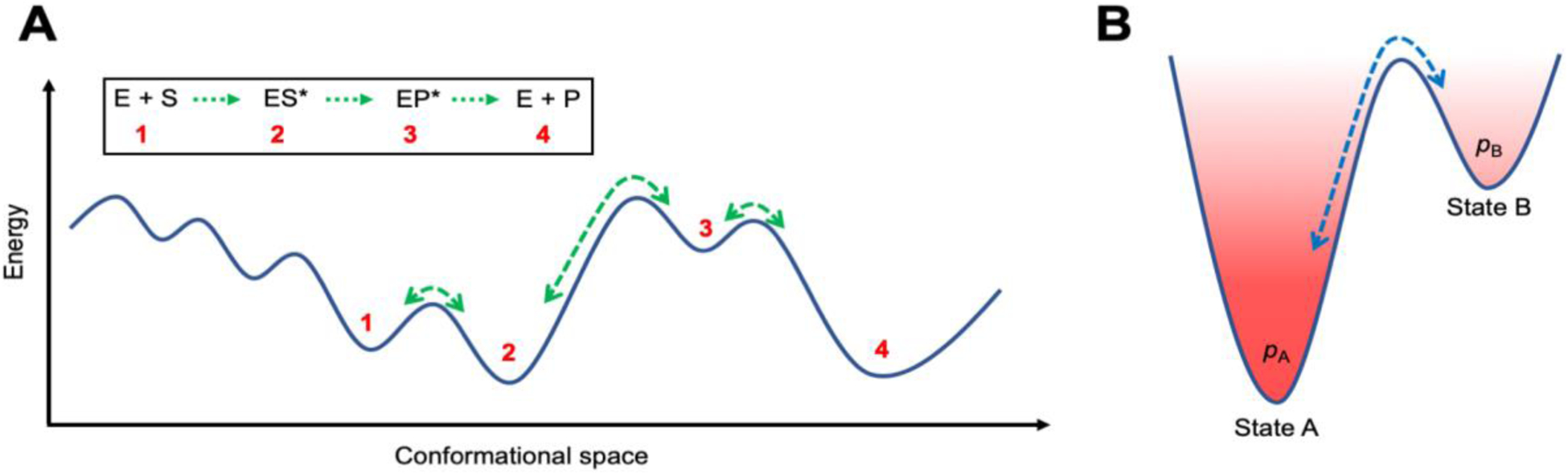
(A) Enzymes do not stay fixed in their ‘native’ structures, but instead undergo conformational motions. Conformational changes experienced by the protein on several time-scales facilitate the different steps of substrate (and/or cofactor) binding, reactant ground state stabilization, chemical step of conversion, and product release along the catalytic cycle of an enzyme. The green arrows over the barriers in the conformational landscape correspond to the rate of transitions between the distinct sub-states. The rates of these conformational transitions are intrinsic properties of an enzyme topology and can be reproducibly measured with appropriate experimental techniques (2, 4). (B) Interconversion between two conformational states A and B. A is the lower energy state compared to state B (excited state), therefore, the conformational populations and associated probability pA will be higher than pB.
