Figure 2. Schematic depiction of the millisecond time frame sampled by TRESI-HDX.
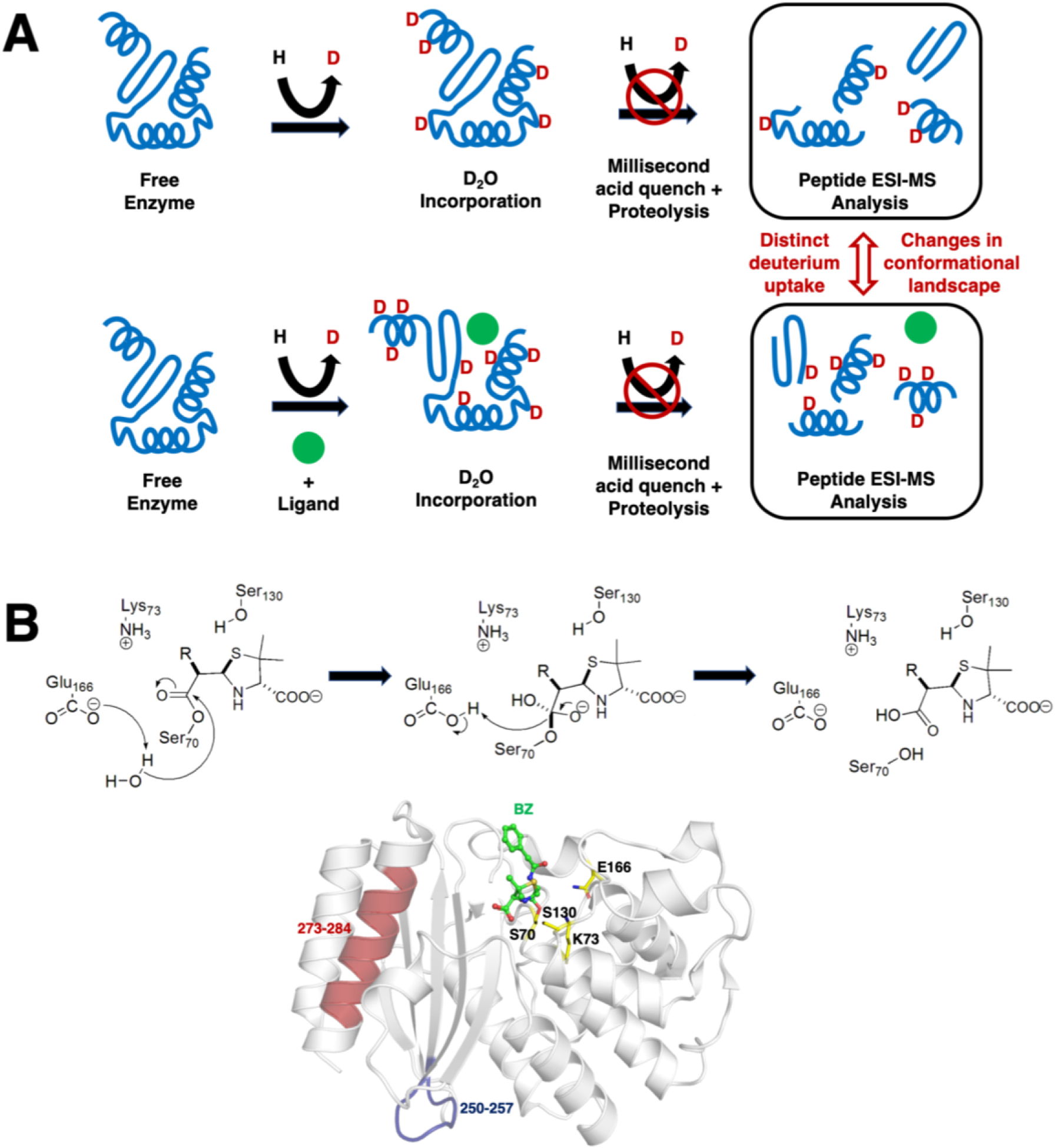
A) The experiment allows comparative analysis of changes in conformational dynamics experienced by a working enzyme during catalysis. The enzyme is first subjected to deuterium incorporation in absence (top) and presence (bottom) of a ligand of interest. To isolate unique dynamic modes associated to specific states of a catalytic cycle, broad ligand diversity can be a significant comparative asset (good or bad substrate affinity, inhibitor, analog, product, etc.). As the protein samples different dynamic modes in its working state, labile/exposed 1H on the structure are replaced by deuterium from the solvent. Deuterium incorporation is further stopped by acid quenching after a given period of time (in this case, milliseconds) and the protein is subjected to proteolysis before peptide ESI-MS analysis. Differences in deuterium uptake effectively allows extraction and comparison of the conformational landscape sampled by the enzyme under the influence of different ligands along the reaction. Figure adapted from ref. (50). B) Catalytic mechanism of deacylation in TEM-1 β-lactamase, whereby Glu166 abstracts a proton from a strictly conserved water molecule to initiate breakdown of the acyl-enzyme intermediate and regenerate the free enzyme. The typical benzylpenicillin (BZ) substrate is illustrated as example, with labeling and mapping of catalytic residues S70, K73, S130 and E166 (yellow carbon coloring) on the crystal structure of the acylated form of TEM-1 with BZ (green carbon coloring). Long-range rigidification of the S4/S5 loop (residues 250–257, in blue) and C-terminal alpha helix of TEM-1 (residues 273–284) have been shown to affect deacylation in TEM-1, possibly through a previously uncharacterized allosteric mechanism (50).
