Figure 6.
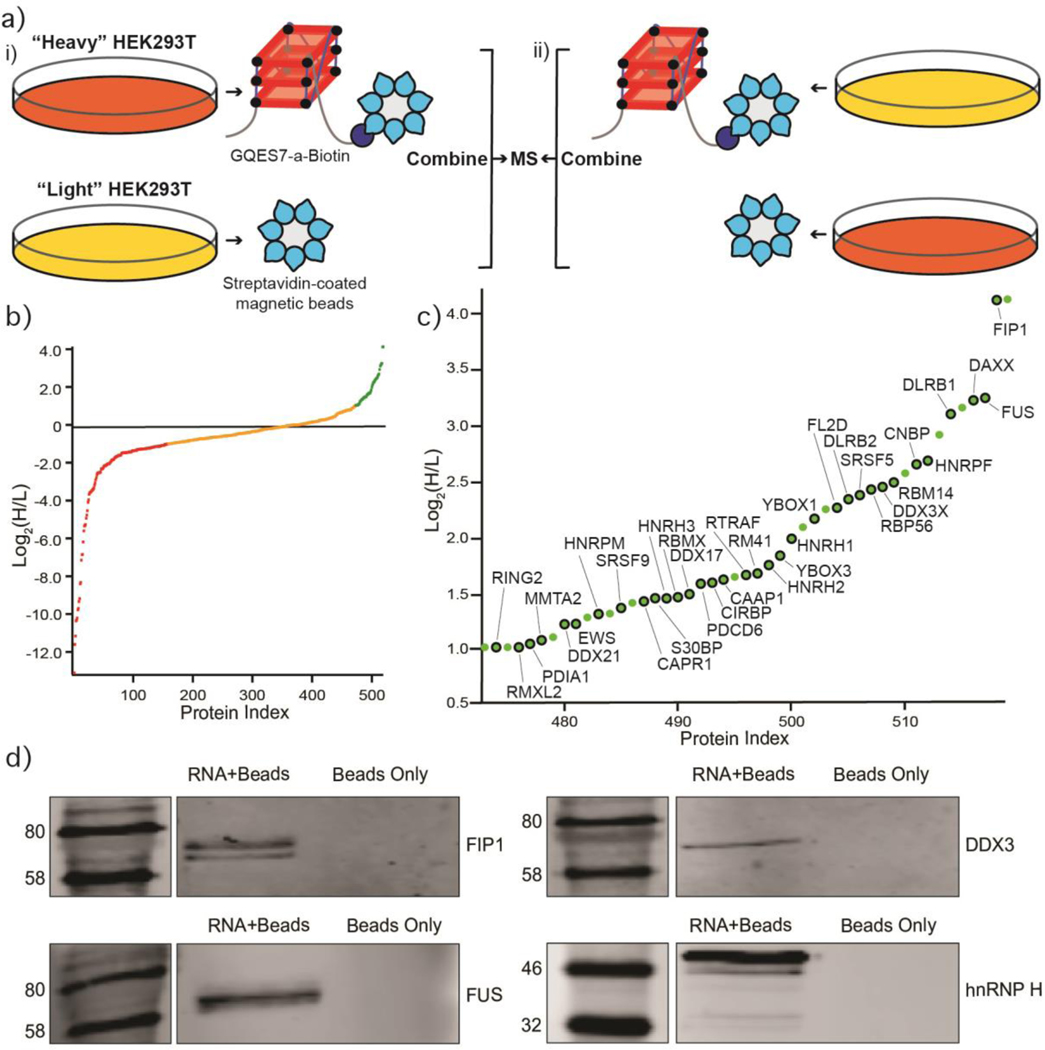
Identification of GQES7-a-binding proteins. a) Scheme of the SILAC experiment. “RNA+beads” samples were combined in HEK293T grown in heavy media (panel i). The “Beads Only” control sample was combined in HEK293T grown in light media. To verify the proteins identified by this method, the experiment was performed using reverse labeling (panel ii). b) Scatter plot representing fold enrichment of the proteins binding to GQES7-a in “Heavy” HEK293T. Color representation indicates specific proteins that bound more tightly to GQES7-a than to the beads (green), to the beads than to GQES7-a (red) or bound to the beads and GQES7-a to a similar extent (orange). c) A close-up view of the green region of the scatter plot represented in panel b. Dots with a black contour are used to indicate proteins that appeared in the green region of the two replicate experiments described in panel a. d) Western blotting analyses of the eluted proteins from the RNA pull-down of HEK293T. All four blotted proteins (FIP1, FUS, DDX3 and hnRNP H) eluted from the GQES7-a sample (RNA+Beads) but not from the control (Beads Only), confirming the SILAC results.