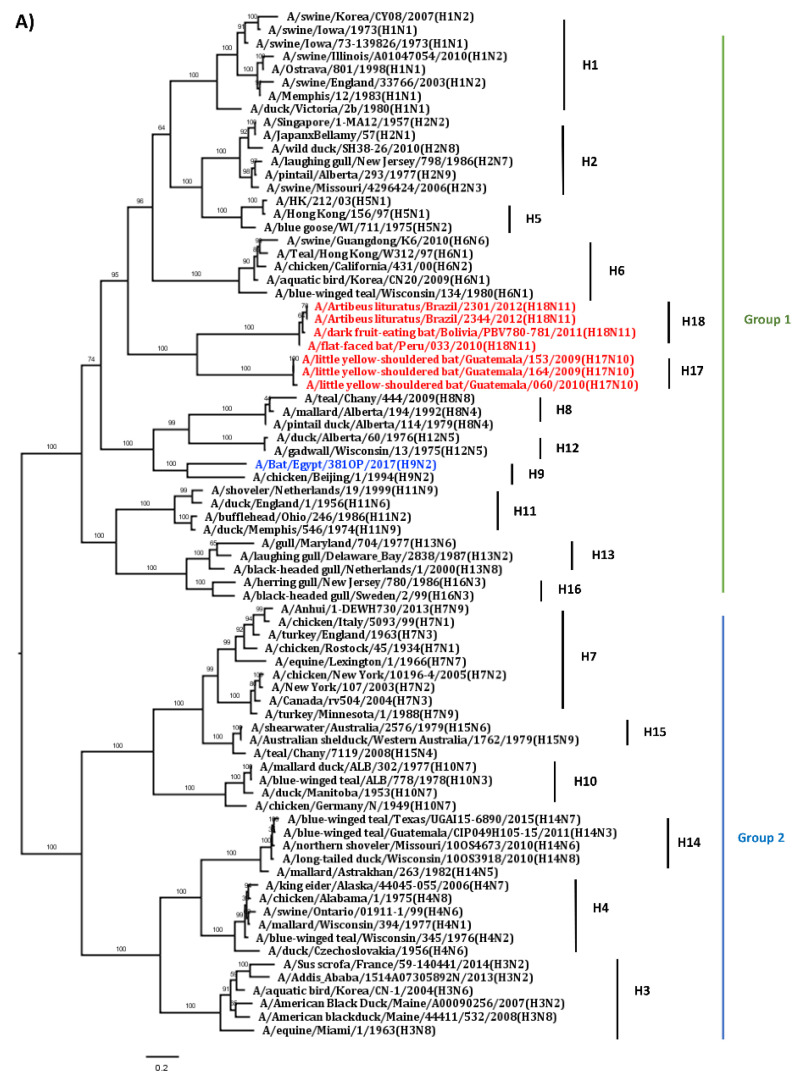
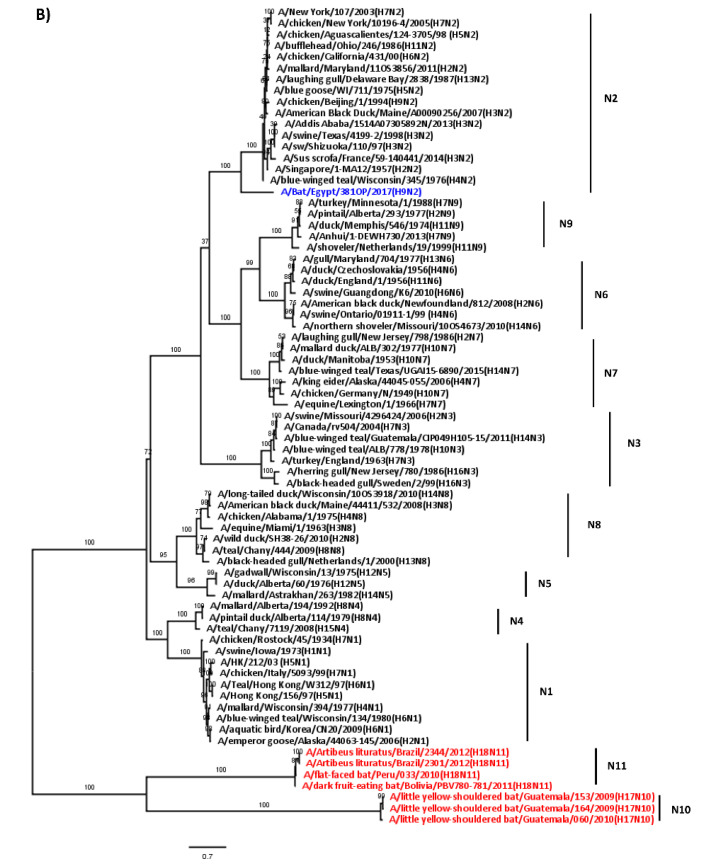
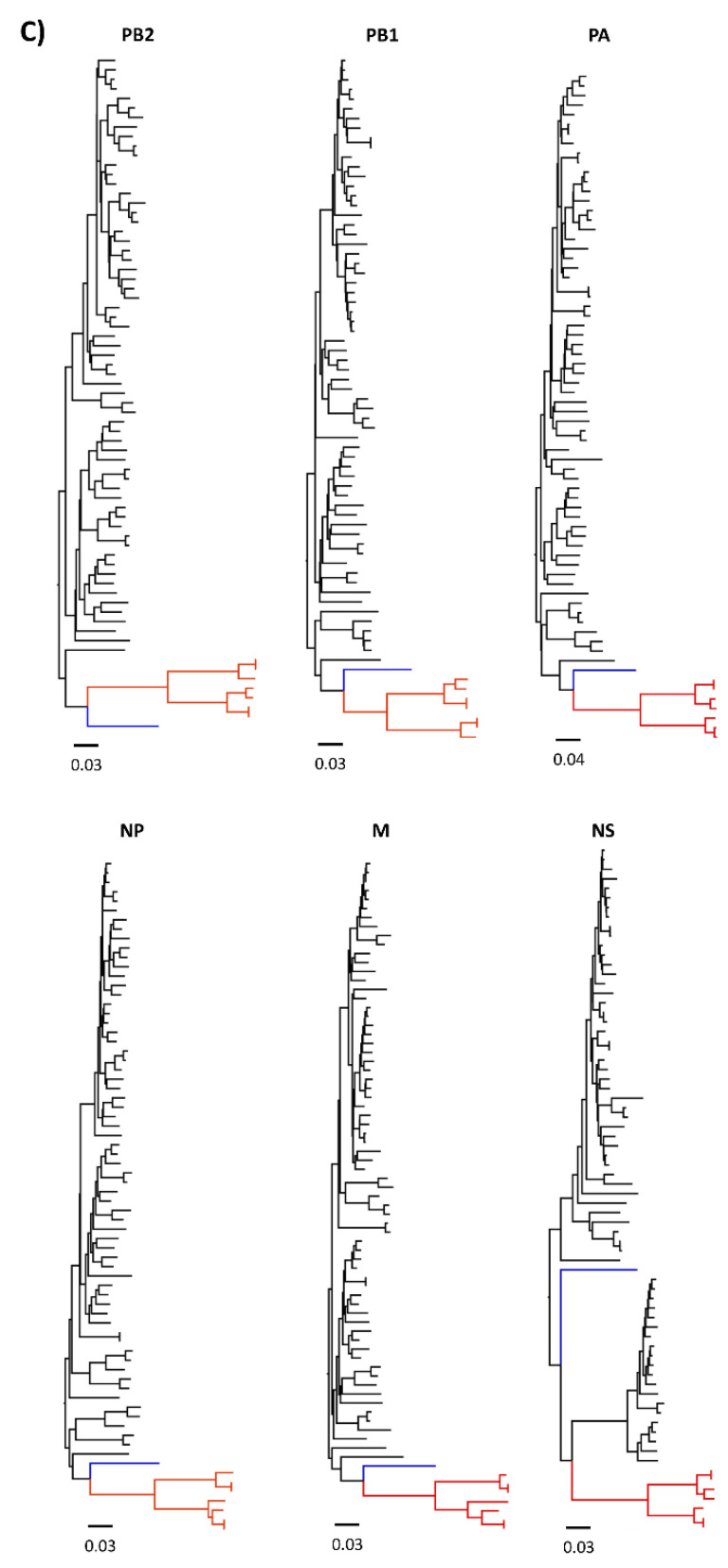
Figure 1.
Phylogenetic trees of bat influenza virus genes. (A) Phylogenetic tree of HA gene. (B) Phylogenetic tree of NA gene. (C) Phylogenetic trees of internal genes. We downloaded 70 full-length genome sequences from different subtypes of conventional influenza A viruses (IAVs) and eight bat influenza viruses, including 3 H17N10, 4 H18N11 and 1 H9N2-like bat viruses from public database. Phylogenetic analyses for each gene were performed based on their full-length gene sequences. The sequences were aligned by the MUSCLE method by MegAlign Pro 17. Phylogenetic relationships among bat HA and NA sequences with those from conventional IAVs were calculated using the maximum likelihood method by MegAlign Pro 17, while for the internal genes determined by using the Neighbor joining method by MegAlign Pro 17. Phylogenetic trees were visualized in FigTree (v1.4.3; http://tree.bio.ed.ac.uk/software/figtree/). Red color represents bat H17N10 and H18N11 viruses; blue color represents A/Bat/Egypt/381OP/2017 (H9N2) virus.