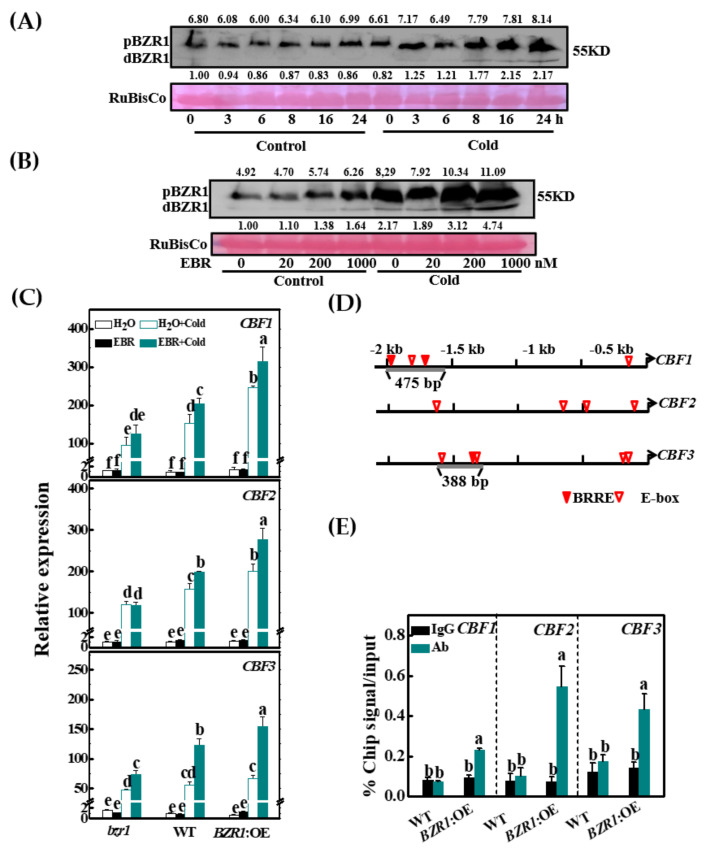
Figure 2.
Cold-/BR-induced BZR1 functions as a transcription regulator for CBF expression. (A) The accumulation of BZR1 protein from tomato leaves exposed to 25 °C (control) or 8 °C (cold) for different periods of time. The values above and below each lane indicate the relative accumulation of pBZR1 and dBZR1, respectively. (B) The effect of 24-epibrassinolide (EBR) on the accumulation of BZR1 protein from tomato leaves under control and cold conditions. Plants were foliar applied with EBR at different concentrations for 24 h and subsequently exposed to cold at 8 °C for another 12 h. In addition, 35S:BZR1-3HA-overexpressing (BZR1:OE) plants were used for analysis for BZR1 protein. (C) The transcripts of CBFs in bzr1 mutant, wild-type (WT), and BZR1:OE transgenic plants exposed to 25 or 8 °C for 8 h; 24 h before cold treatment, the plants were foliar applied with 200 nM EBR or distilled water as the control. (D) BRRE and E-boxes in the tomato CBF promoter sequences. Numbering is from predicted transcriptional start sites. (E) ChIP-qPCR analysis of BZR1 binding to the promoters of CBF1, CBF2, and CBF3 in tomato. Input chromatin was isolated from leaves exposed to 8 °C for 8 h. CBF promoter fragments were immunoprecipitated with an anti-HA antibody. Mouse IgG was used in control reactions. Input- and ChIP-DNA samples were quantified by qRT-PCR. The ChIP data are shown as % input DNA. Data are the means of three biological replicates (±SD) shown by vertical error bars. Different letters indicate significant differences according to Tukey’s test at 0.05% level.