Figure 5.
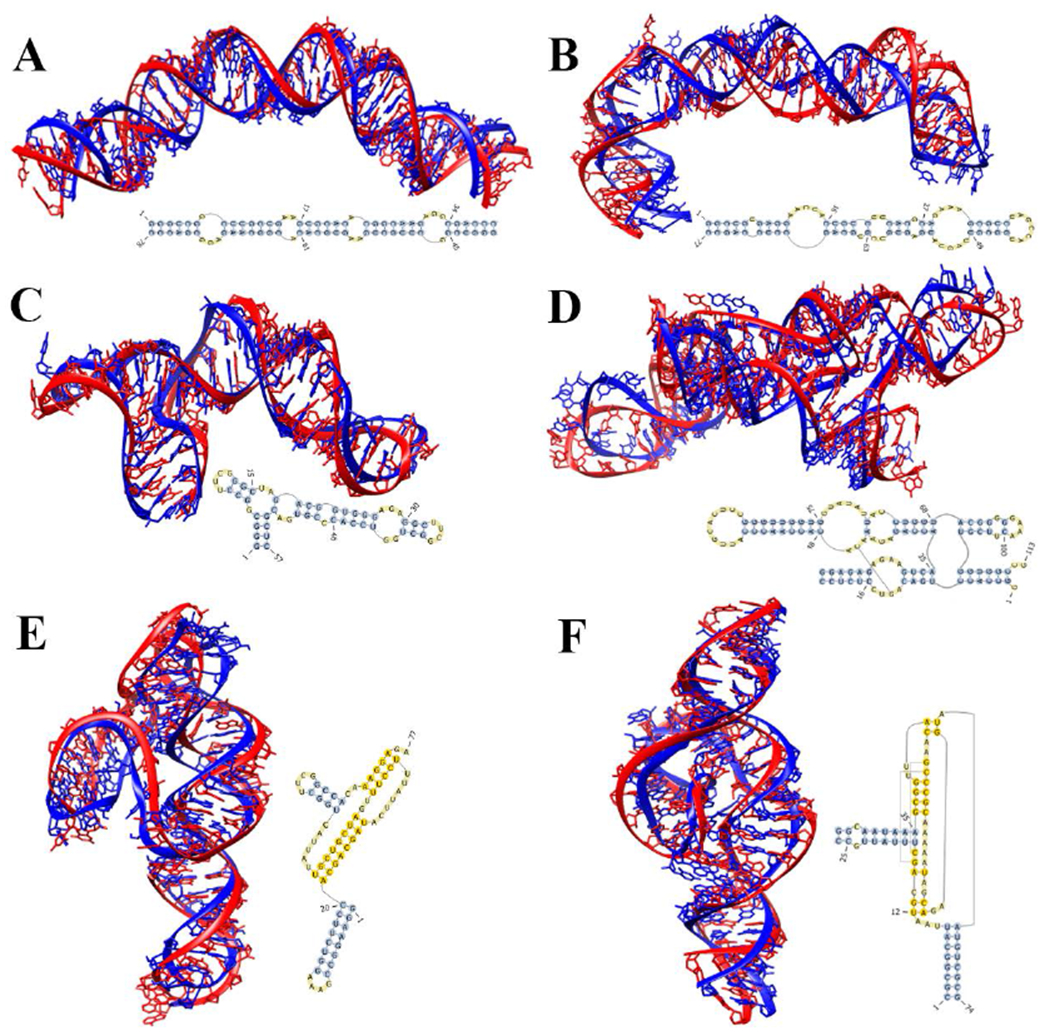
Predicted 3D structures by IsRNA1 model for several representative RNA molecules. (A) 78-nt HIV-1 dimerization initiation site in the extended-duplex dimer77 (PDB ID: 2d1a), (B) 77-nt HCV IRES domain II78 (PDB ID: 1p5p), (C) 57-nt rRNA fragment in the S15-rRNA complex79 (PDB ID: 1dk1), (D) 21-nt RNA substrate and 92-nt RNA hairpin ribozyme complex80 (PDB ID: 1m5o), (E) 77-nt PreQ1-ll riboswitch83 (PDB ID: 5d5l), and (F) 74-nt twister ribozyme84 (PDB ID: 4qjh). The predicted structures of lowest RMSD from the top 3 predictions given by IsRNA1 (in red) are superposed onto the native structures (in blue). Secondary structures used to impose constraints on IsRNA1 modeling are also. RMSDs over all heavy atoms are 4.12 Å, 6.89 Å, 5.37 Å, 5.99 Å, 6.54 Å, and 4.20 Å, respectively.
