Fig. 1. The computational approach used to resolve the structures of nucleic acid duplexes.
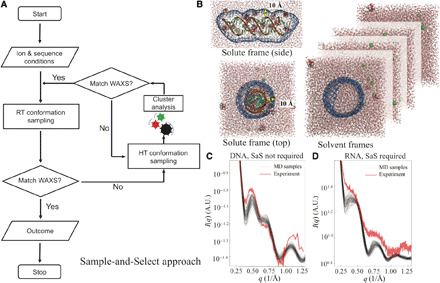
(A) SaS approach with enhanced sampling at higher temperature and selection based on agreement with experimental WAXS profiles. The feedback loop is triggered when the customized metric in Eq. 6 of all the sampled conformations is greater than 10.0. (B) Illustration of the molecular envelope constructed from a 3D probability density isosurface 10 Å from the RNA surface to encompass the solvent/ion shell. The same envelope was applied to solvent systems to compute the scattering amplitudes to be subtracted. (C) The WAXS profiles from experiment (red) and MD simulations with starting B-form conformation (black) of DNA duplex. Here, the sampling captures features of the experimental data. The selection feedback is not necessary. (D) Same as (C) for an RNA duplex. Here, none of the WAXS profiles from MD simulations (black) agree with experimental data (red). The SaS approach is required to enhance sampling to identify a plausible starting conformation.
