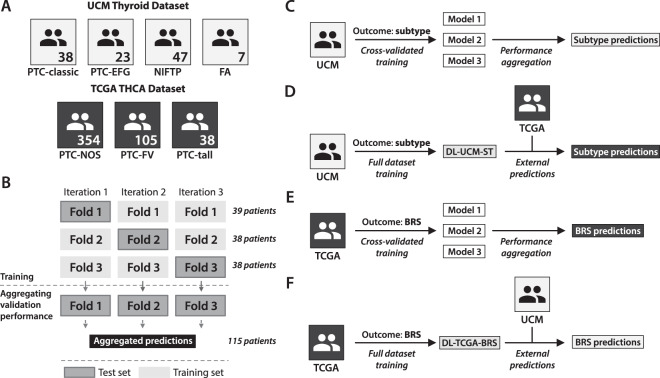
Fig. 1. Deep learning experimental strategy.
a Distribution of thyroid neoplasm subtypes within the institutional (University of Chicago Medicine, UCM) and TCGA datasets. The institutional dataset contained a total of 115 slides across four subtypes, as detailed here. The TCGA thyroid cohort (THCA) contained 497 slides across three subtypes: papillary thyroid carcinoma, not otherwise specified (PTC-NOS), papillary thyroid carcinoma, follicular variant (PTC-FV), and papillary thyroid carcinoma, columnar cell variant (PTC-tall). As the annotated subtypes in TCGA use an older naming convention before the recognition of NIFTP, these annotations cannot be directly compared between datasets. b Cross-validation plan for training a deep learning model. A deep learning model was first trained on the institutional dataset to predict tumor subtype, using threefold cross-validation as illustrated here. Patients were separated into three categories of 39, 38, and 38 patients per category, with annotated subtypes balanced between categories. For each of three iterations, two-thirds of the dataset was used for training and one-third was used for validation to generate predictions. Predictions in each of the left-out validation sets were aggregated across the three k-fold iterations in order to generate the reported cross-validation performance statistics. c Overview of the deep learning training strategy used to generate cross-validated subtype predictions on the institutional dataset, as also described in (b). d After cross-validation was completed, a final model (DL-UCM-ST) was trained across the entire institutional dataset to predict subtype. This model was applied to the TCGA dataset, for which subtype predictions were generated. e Deep learning models were trained on the TCGA dataset to predict BRS, using cross-validation as described in (b). f After cross-validation was completed, a final model (DL-TCGA-BRS) was trained across all TCGA slides to predict BRS. This model was then used to generate BRS predictions on the institutional dataset.