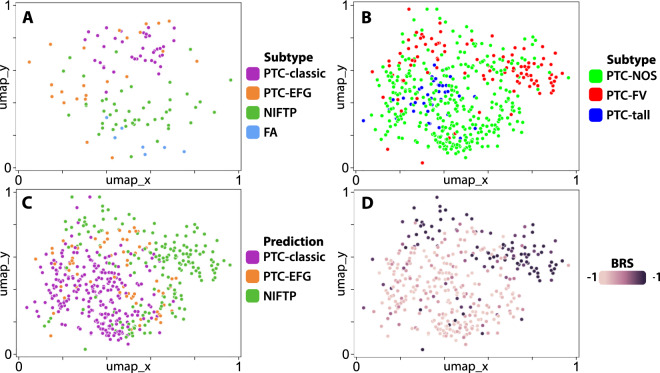
Fig. 2. UMAPs of post-convolution activations generated by deep learning models trained to predict tumor subtype.
a An Xception-based deep learning model was trained on 115 slides from an institutional database of thyroid cancer to predict tumor subtype (DL-UCM-ST). Post-convolution layer activations were calculated on all tiles and mapped with UMAP. For each slide, the tile nearest to centroid was plotted and labeled according to its tumor subtype. NIFTP slides can be seen to cluster distinctly from the PTC subtypes, clustering near follicular adenomas. Several PTC-EFGs cluster near PTC-classics, but approximately half are found between the NIFTP and PTC-classic clusters. b The DL-UCM-ST model was used to generate slide-level predictions in the TCGA thyroid cohort. Slides with follicular-pattern cluster together, but demonstrate overlap with slides labeled as “Papillary thyroid carcinoma, not otherwise specified” (PTC-NOS). c Same as (b), with slides labeled according to their predicted subclass. Many of the true PTC-EFG tumors are predicted to be NIFTPs, as can be seen by comparing this plot to (b). Tumors which are predicted to be NIFTP cluster distinctly from those predicted to be PTC-classic. Many of the tumors falling on the border of these two clusters are predicted to be PTC-EFGs. d Same as (b), with slides labeled according to their BRS, as calculated by Agarwal using a 71-gene mRNA expression signature [1]. Tumors with BRS > 0 are RAS-like, and BRS < 0 indicate BRAFV600E-like phenotype. Tumors with high BRS, and thus RAS-like phenotype, are highly associated with the cluster of tumors predicted to be NIFTPs, as can be appreciated by comparing this plot with C. The majority of BRAFV600E-like tumors are predicted to be either PTC-classics or PTC-EFGs.