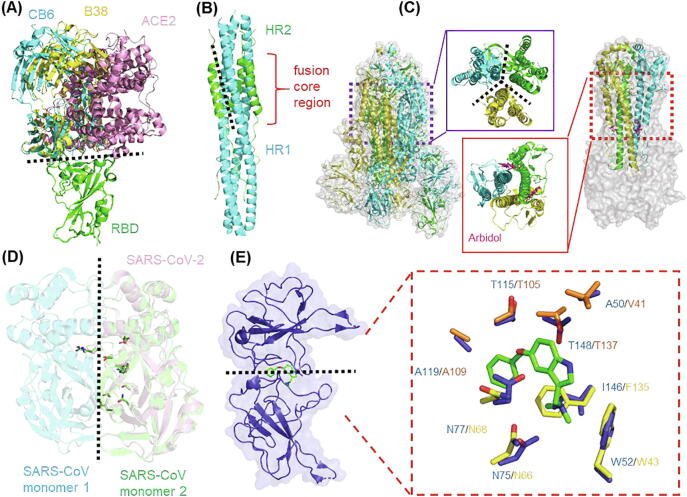
Fig. 3.
Main strategies for PPI modulator design against SARS-CoV-2. (A) Hot-spot for PPI inhibitor design against RBD-ACE2 complex. Structures of SARS-CoV-2 S protein RBD in complex with ACE2 (PDB: 6LZG), CB6 (PDB: 7C01) and B38 (PDB: 7BZ5) complex are shown in cartoon. The RBDs of each structure are aligned to show the PPI suitable for modulator design. (B) Hot-spot for PPI inhibitor design against fusion core region of S protein. SARS-CoV-2 6-HB structure is shown in cartoon with HR1 and HR2, colored in green and cyan, respectively (PDB: 6LXT). (C) Hot-spot for PPI modulator design against S protein trimerization. (Left) Structure of SARS-CoV-2 S protein trimer (PDB: 6VSB). The trimeric interface is enlarged in the middle. (Right) Structure of the influenza HA in complex with arbidol (PDB: 5T6N). The arbidol target site of the trimeric interface is enlarged in the middle. (D) Hot-spot for PPI inhibitor design against intra-dimer of 3CLpro. The structure of 3CLpro of SARS-CoV-2 (PDB:6Y2E) is aligned with that of SARS-CoV (PDB: 1UK4). The key residues involved in dimerization are shown in stick representation. (E) Hot-spot for PPI stabilizer design against N-NTD. The structure of SARS-CoV-2 N-NTD (PDB: 6M3M) is aligned to P3: MERS CoV N-NTD complex (PDB: 6KL6), the interacting residues are shown in sticks and highlighted in right box. The residues of SARS-CoV-2 and MERS CoV N-NTD are shown in orange and yellow, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)