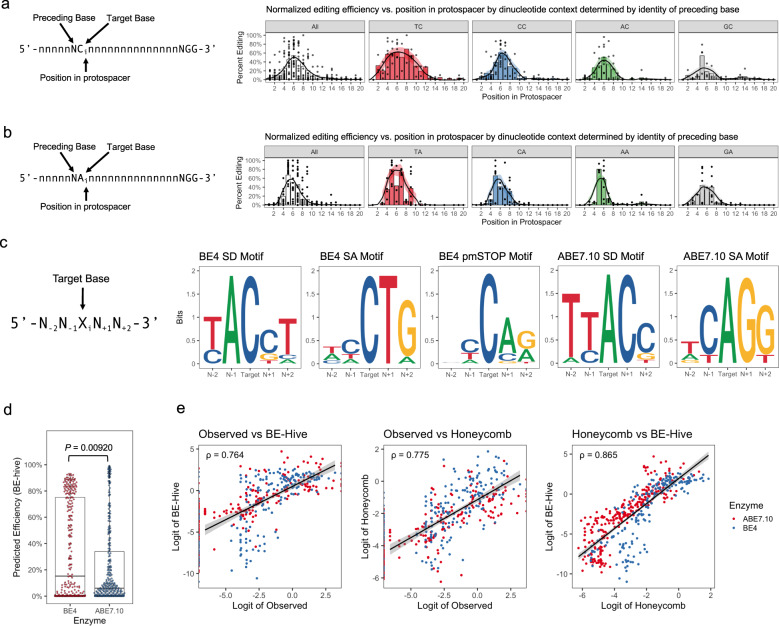
Fig. 4. Context dependencies of base editors and BE-splice target motifs.
a Dinucleotide context dependencies of rAPOBEC1-BE4. Dinucleotide context is defined by the identity of the target base, and the identity of the base immediately preceding the target. Results normalized and aggregated across published results and our own work. Smoothed distributions generated using LOESS regression with span = 0.5. b Preceding dinucleotide context dependencies of TadAWT-TadAEvo-ABE7.10. Results normalized and aggregated across published results and our own work. Smoothed distributions generated using LOESS regression with span = 0.45. N = 6 papers, 102 guides, and 447 edits in the analysis. c Logo plots of the pentanucleotide motif for each enzyme and target motif combination in this work. The pentanucleotide motif is oriented with respect to the sgRNA protospacer, independent of how a protospacer is oriented with respect to the direction of gene transcription. The heights of bases are proportional to the prevalence of the base at that position in the target site. d Boxplot of BE-Hive predicted editing efficiencies between BE4 and ABE7.10 sgRNAs. Boxplot center lines represent the median, box limits represent the upper and lower quartiles, and whiskers define the 1.5× interquartile range. Analyzed by two-sided Wilcoxon rank sum test due to the non-normal distribution of data. N = 275 BE4 edits, and 342 ABE7.10 edits. e Comparisons between observed values in meta-analysis and BE-Hive or Honeycomb predicted scorings. Data plotted on a logit scale to better observe relationship of data. Spearman’s rank correlation coefficient (ρ) is shown. Trend line is linear model line of best fit, grey shading is 95% CI of the mean. Source data are available in the Source Data file.