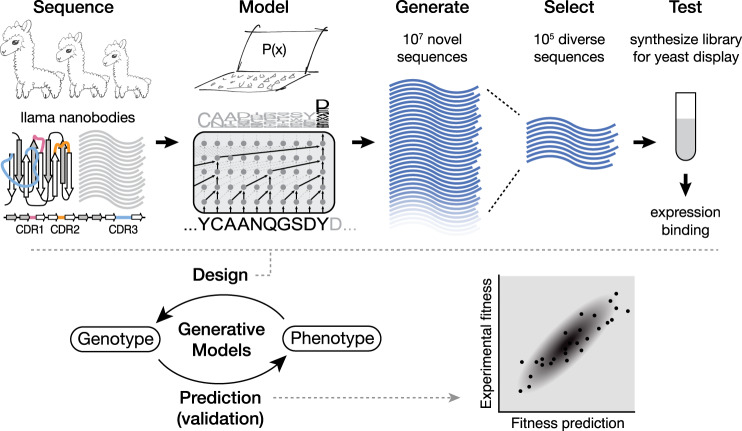
Fig. 1. Autoregressive models of biological sequences can learn the genotype-phenotype map for both prediction and design.
From natural sequences (gray) in a naïve llama repertoire57, the autoregressive model can learn functional constraints by predicting the likelihood of each residue in the sequence conditioned on preceding residues. Nanobodies have three highly variable complementarity determining regions (CDR1, CDR2, and CDR3). We then use these constraints to generate millions of novel nanobody sequences (blue)—as many can be generated as desired. Of these designed sequences we select hundreds of thousands of diverse sequences, synthesize a library, and screen for expression and binding. We also validate the model on mutation effect prediction tasks of deep mutational scans including the effects of multiple insertions and deletions, and the thermostabilities of highly variable nanobody sequences.