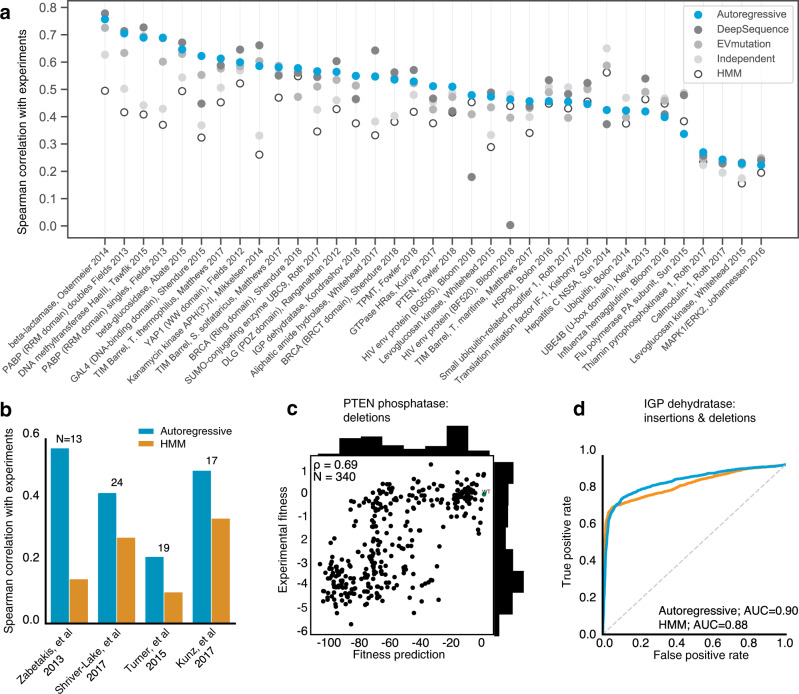
Fig. 2. Validation of the autoregressive model in learning the genotype to phenotype map.
a Even without using alignments, the autoregressive model (blue) can competitively match mutation effect prediction accuracies of state-of-art alignment-dependent models, such as conservation (light gray), evolutionary couplings (gray), and DeepSequence (dark gray)30. In addition, the mutation effect prediction accuracies improve upon hidden Markov model74 (HMM, white) accuracies. Without using alignments, the autoregressive model matches alignment-dependent state-of-art missense mutation effect prediction (DeepSequence) for 40 different deep mutational scan experiments. Three datasets show significant improvement with the autoregressive model: HIV env (BF520), HIV env (BG505), and Gal4 DNA-binding domain. b The autoregressive model (blue) can learn from natural sequence repertoires of llama nanobodies to predict the thermostability of llama nanobody sequences with variation in the framework and complementarity determining regions with greater accuracy than HMMs (orange). The number of llama nanobody sequences from each study is shown above each pair of bars. c Fitness predictions for single deletions in PTEN phosphatase compared with measured experimental fitness is accurate, with a Spearman correlation of 0.69. d Accurate prediction of binary fitness for IGP dehydratase with a range of insertions, deletions, and missense mutations of the autoregressive model (blue), higher than HMM (orange).