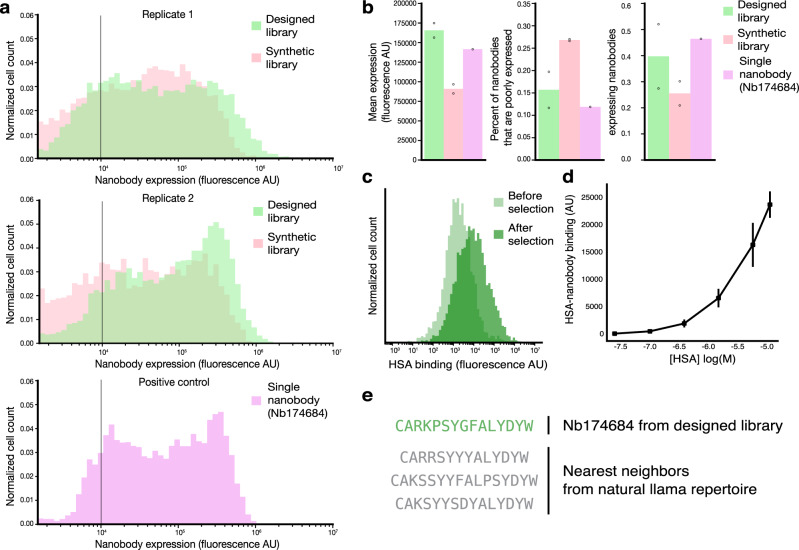
Fig. 4. The designed library contains stable and functional nanobody sequences that are well expressed and can bind target antigens.
a Fluorescence distributions of cells expressing nanobodies comparing the synthetic combinatorial library (pink) and our designed library (green) in two biological replicate experiments as well as a control experiment of a single, well-expressed nanobody clone (Nb174684, purple). The distributions of the designed library are consistently right-shifted compared to the combinatorial library and resemble the control nanobody. b Compared to the combinatorial library, the designed library has almost double the mean expression level (166,193 AU compared to 92,183 AU), nearly half the fraction of poorly expressed nanobodies (of cells expressing nanobodies) (15.4% compared to 25.7% of clones with less than 10,000 AU indicated as a gray bar in panel (a)), and one and a half times the fraction of total cells that express nanobodies (39.6% compared to 25.1%). The thresholds for determining the proportion of total cells expressing nanobodies were found by identifying the local minima on the distributions and are displayed in Supplementary Fig. 10. Values displayed on the bar graphs are means of two biological replicates for the two libraries and one replicate for the control experiment of the single nanobody clone. Replicate measurements are displayed as dots for the two library experiments. c Fluorescence distributions of nanobodies bound to HSA show a rightward shift after screening and selection, indicating a successful enrichment of binders to the target antigen. d On-yeast binding assay of Nb.174684, an HSA binder identified from the designed library with moderate binding affinity. The means of HSA binding (AU) of three replicates are shown and error bars represent standard deviations in measurements at each concentration of HSA. e CDR3 sequence of binder Nb.174684 and the sequences of the nearest neighbors from the natural llama repertoire that was used to train the autoregressive model.