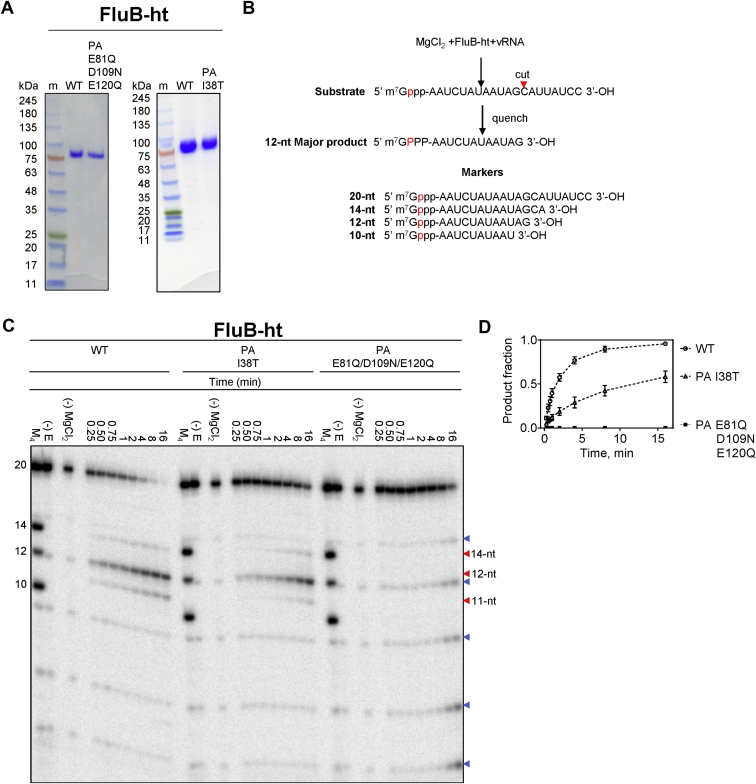
Figure 2.
Purification and biochemical characterization of the Influenza B polymerase heterotrimer (FluB-ht).A, SDS-PAGE migration patterns of the purified WT FluB-ht, baloxavir resistance mutant (PA I38T), or endonuclease-deficient mutant (PA E81Q/D109N/E120Q). Enzyme preparations were stained with Coomassie brilliant blue R-250. The size of the molecular weight markers (m) is indicated in kDa to the left of the gel. The band above the 75 kDa marker corresponds to full-length FluB-ht containing PA, PB1, and PB2 subunits. The identity of each of the subunits was confirmed via LC-MS/MS. B, schematic representation of the nuclease reaction with the position of the radiolabel highlighted in red. The sequences of the 20, 14, 12, and 10-nt markers are highlighted (Lane M4). C, endonuclease activity of WT, PA I38T, or endonuclease-deficient (PA E81Q/D109N/E120Q) FluB-ht on PAGE. M4 indicates the migration patterns of an equimolar mixture of 5’- m7G capped 20-nt, 14-nt, 12-nt, and 10-nt oligos here utilized as molecular weight markers (–) E indicates the migration pattern of 5’capped 20-nt substrate in the absence of enzyme. (–) MgCl2 indicates the results of the reaction after 16 min in the absence of Mg2+. Blue arrows indicate hydrolysis products while red arrows indicate the position of bona fide endonucleolytic cleavages. D, graphical representation of data shown in A. Data points are connected using a dotted line to illustrate the progress of the reaction. Product fraction refers to the ratio of the signal produced by the 5’ capped 11- and 12-nt products to the sum of these products plus the remaining substrate. The 14-nt product was not quantified as its contribution to overall signal was negligible. Error bars represent the standard deviation of at least three independent experiments. All experiments were performed under the following final conditions: 55 nM FluB-ht, 100 nM 20-nt substrate, 30 mM Tris-pH 7.5, 25 mM NaCl, 1.7 μM vRNA, and 5 mM MgCl2.