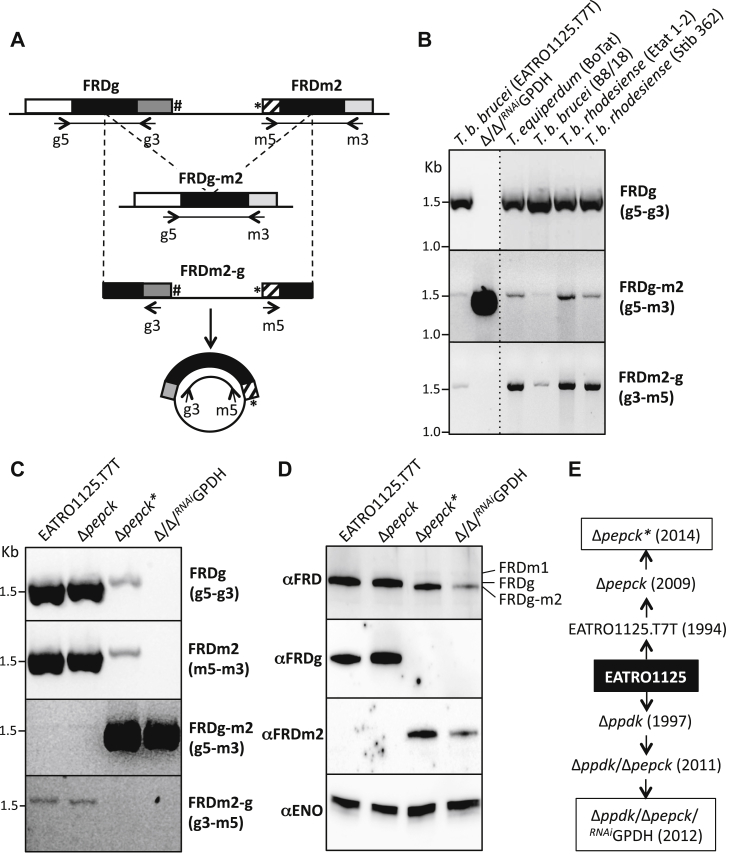
Figure 4.
PCR analysis of the recombination event in the FRDg/FRDm2 locus. The PCR strategy developed to detect a recombination event within the FRDg/FRDm2 locus is described in (A). This schematic representation shows the DNA recombination event leading to the deletion of the FRDm2-g fragment, which can be circularized by ligation, as well as the position of the primers (arrows) designed to amplify the FRD domains of FRDg (g5-g3; 1583 bp), FRDm2 (m5-m3; 1611 bp), and the chimeric FRDg-m2 (g5-m3; 1577 bp), as well as the recircularized deleted FRDm2-g fragment (g3-m5; 1608 bp). See Figure 2B legend for the color code of the genes. B, shows a PCR analysis of the EATRO1125.T7T parental and Δppdk/Δpepck/RNAiGPDH cell lines, as well as four additional African trypanosome strains, using 100 ng of genomic DNA (10 μg ml−1). In panelsC and D, a comparative analysis of the parental EATRO1125.T7T, Δpepck∗, Δpepck, and Δppdk/Δpepck/RNAiGPDH cell lines is presented, using the PCR approach described in panel A (C) and the western blot analysis using antibodies specific for enolase (αENO), FRDg (αFRDg), FRDm2 (αFRDm2), or the three T. brucei FRD isoforms (αFRD) (D). The history of the Δppdk/Δpepck/RNAiGPDH and Δpepck∗ cell lines (boxed), which have selected the recombinant FRDg/FRDm2 locus, is shown in (E). The Δpepck cell line obtained in 2009 contains the parental FRDg/FRDm2 locus, which became recombined later after long-term in vitro culture (Δpepck∗ cell line, 2014).