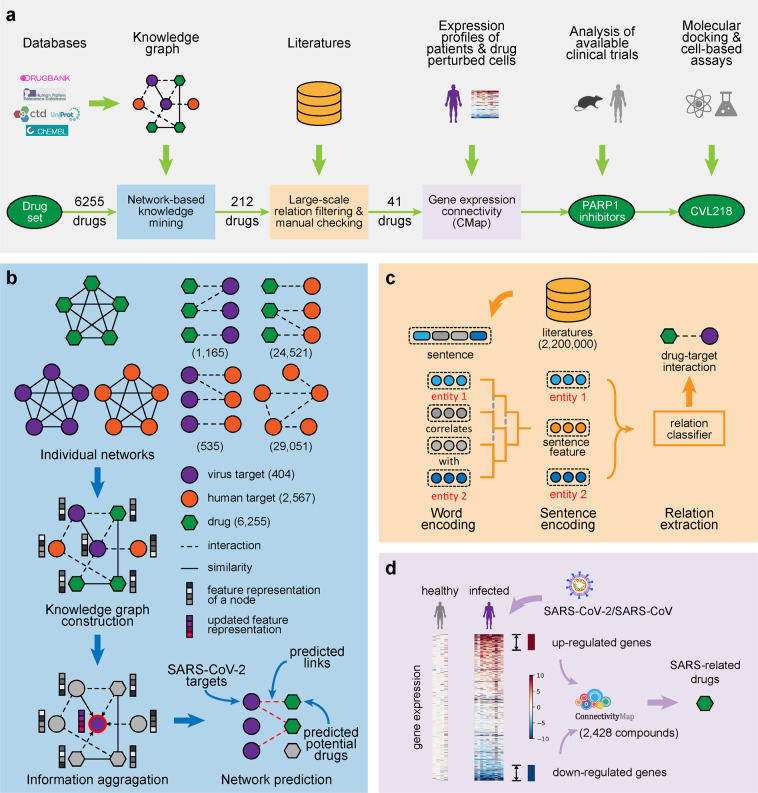
Fig. 1.
Schematic illustration of our integrative drug repositioning pipeline for discovering the potential drugs to treat the COVID-19 disease. a The overview of our drug screening pipeline. The initial drug set for screening contains 6255 drug candidates, mainly including 1786 approved, 1125 investigational, and 3290 experimental drugs. The number of drug candidates after each filtering step is also shown. Knowledge graph: a network containing entities (e.g., drugs and targets) and their relations. b The network-based knowledge mining module. Seven individual networks containing three types of nodes (i.e., drugs, human targets and virus targets) and the corresponding edges describing their interactions, associations or similarities are first constructed based on the known chemical structures, protein sequences and relations derived from publically available databases. Then a deep learning based method, which learns and updates the feature representation of each node through information aggregation, is used to predict the potential drug candidates against a specific coronavirus. c The automated relation extraction module. The structure of each sentence from the literature texts is first learned from the encoded word features using the Gumbel tree gated recurrent unit technique.4,96 Then the learned sequence structures as well as the corresponding encoded word features are fed into a relation classifier to automatically extract the relations between two entities from large-scale documents in the literature. d The connectivity map (CMap) analysis module. The transcriptome profiles of the peripheral blood mononuclear cell (PBMC) or the bronchoalveolar lavage fluid (BALF) samples from the SARS-CoV-2 or SARS-CoV infected patients and healthy persons are compared to derive the query gene expression signatures, which are then correlated to the drug-perturbed cellular expression profiles in the connectivity map5 to filter out the anti-SARS-CoV-2 drug candidates. For transcriptome data of SARS-CoV-2 infected cells, PBMC samples were provided by three patients and three healthy volunteers, and BALF samples were collected from three patients and two healthy volunteers. For transcriptome data of SARS-CoV infected cells, PBMC samples from ten patients and four healthy volunteers were included in our analysis