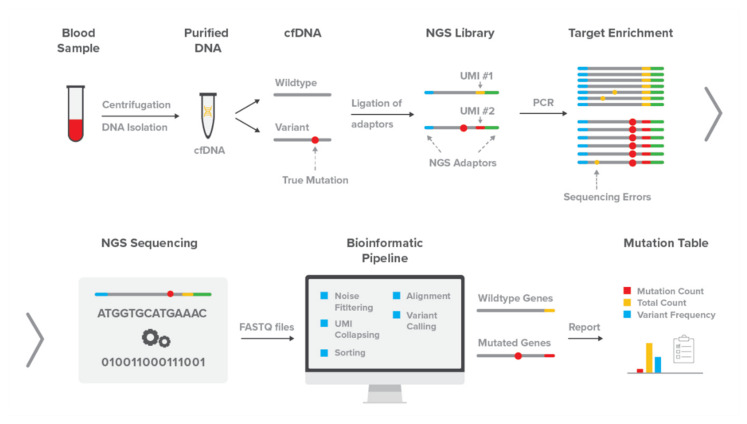
Figure 1.
Overview of the DEEPGENTM assay workflow. Blood samples from liquid biopsies or reference materials are used to isolate cell-free DNA (cfDNA). Unique molecular identifiers (UMI) are ligated to individual cfDNA strands, followed by an enrichment of cancer-relevant sequences using a customized primer panel. The enriched NGS libraries are subsequently sequenced on a Illumina NovaSeq 6000 and analyzed with the DEEPGENTM bioinformatics pipeline. The output is a mutation table that contains the variant frequency for all targeted locations.