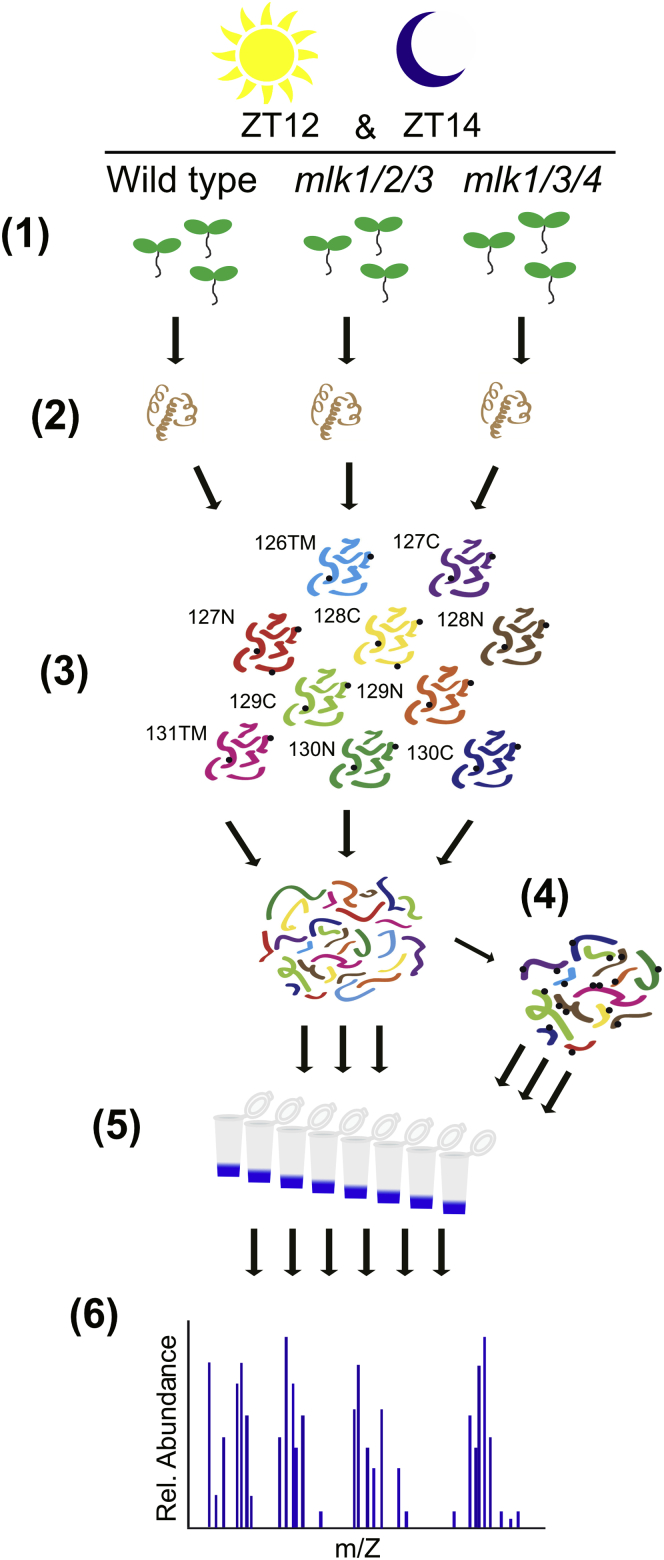
Fig. 1.
Schematic representation of the quantitative proteomics workflow. Tissue samples were collected at ZT12 and ZT14 from WT and mutant seedlings entrained with a 12L:12D light/dark cycle (1). Total protein was extracted and digested (2). Following TMT10plex isobaric labeling (3), samples were subjected to high pH reversed phase prefractionation (5). Phosphopeptides were enriched using a TiO2-based method (4). Both phosphopeptide-enriched and global samples were analyzed by LC-MS/MS (6). ZT 12, Zeitgeber 12; ZT 14, Zeitgeber 14.