Figure 1. AHR modulates pathways relevant to vascular disease and induces chromatin accessibility in HCASMC.
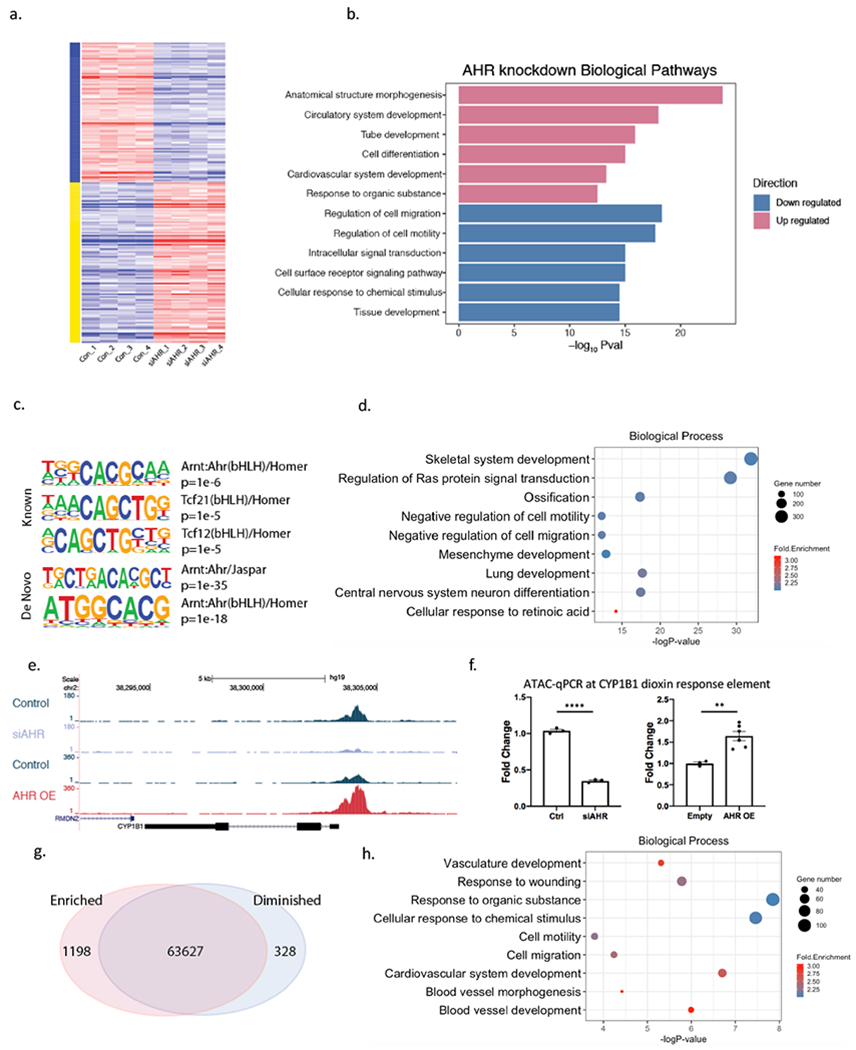
(a) Heatmap of differentially expressed genes in AHR knockdown RNA-Seq data in HCASMC show a significant impact of AHR modulation on the transcriptome. (b) Top enriched biological pathways from differentially expressed genes of AHR knockdown. (c) AHR ChIP-Seq peaks localize to AHR and TCF21 motifs with HOMER known and de novo analyses. (d) DAVID Gene Ontology analysis of AHR target genes identified by GREAT. (e) An ATAC-seq peak is visualized at the promoter region of CYP1B1, where the peak is diminished with knockdown and increased with AHR overexpression and (f) was confirmed with ATAC-qPCR (AHR knockdown by siRNA p<0.0001 (left), AHR overexpression p=0.0048 (right)). ****P < 0.0001, **P < 0.01. (g) AHR overexpression leads to an overall increase in open-chromatin regions. (h) DAVID Gene Ontology analysis of genes in the regions of increased chromatin accessibility signatures from AHR overexpression.
