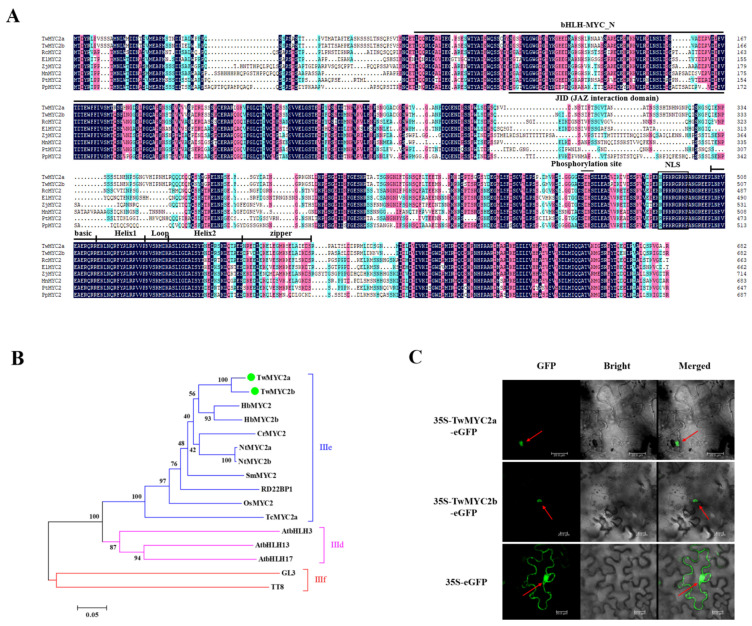
Figure 1.
Multiple sequence alignment, phylogenetic analysis, and subcellular localization analyses of TwMYC2a/b. (A) Multiple sequence alignment of the TwMYC2a/b protein sequences with their homologous MYC2 proteins from different plant species: Ricinus communis RcMYC2 (XP_002519814.1), Euphorbia lathyris ElMYC2 (AVA18027.1), Ziziphus jujuba ZjMYC2 (XP_015896923.1), Morus notabilis MnMYC2 (XP_010104300.1), Populus tomentosa PtMYC2 (AZQ19203.1), and Prunus persica PpMYC2 (XP_020419289.1). The bHLH-MYC-N domain, bHLH-leucine zipper domain, and phosphorylation site were marked with an overline. JAZ interaction domain was marked with an underline. The nuclear localization signal (NLS) was represented by a solid box. (B) Phylogenetic analysis of TwMYC2a/b and other clades IIId/IIIe/IIIf bHLHs. Numbers at nodes represent bootstrap values (based on 1000 resamplings). The scale bar indicates the evolutionary distance. TwMYC2a/b are represented by large green dots. Accession NO. in GenBank: AtMYC2 (also called RD22BP1, BAA25078), AtbHLH3 (AAL55710), AtbHLH13 (Q9LNJ5), AtbHLH17 (Q9ZPY8), GL3 (NP_193864), TT8 (OAO98324), TcMYC2a (ATY38591), OsMYC2 (0336P5), SmMYC2 (AO09733), NtMYC2a (ADU60100), NtNYC2b (ADU60101), CrMYC2 (AQ14332), HbMYC2 (AJC01627), and HbMYC2b (XP_021664832). (C) Subcellular localization analyses of the TwMYC2a/b proteins in tobacco (Nicotiana benthamiana). TwMYC2a/b fused in-frame with an enhanced green fluorescent protein (eGFP) was transiently expressed in tobacco leaf cells. The empty vector 35S::eGFP was used as a control. The red arrows indicate the location of the nucleus. Scale bars are 20 μm.