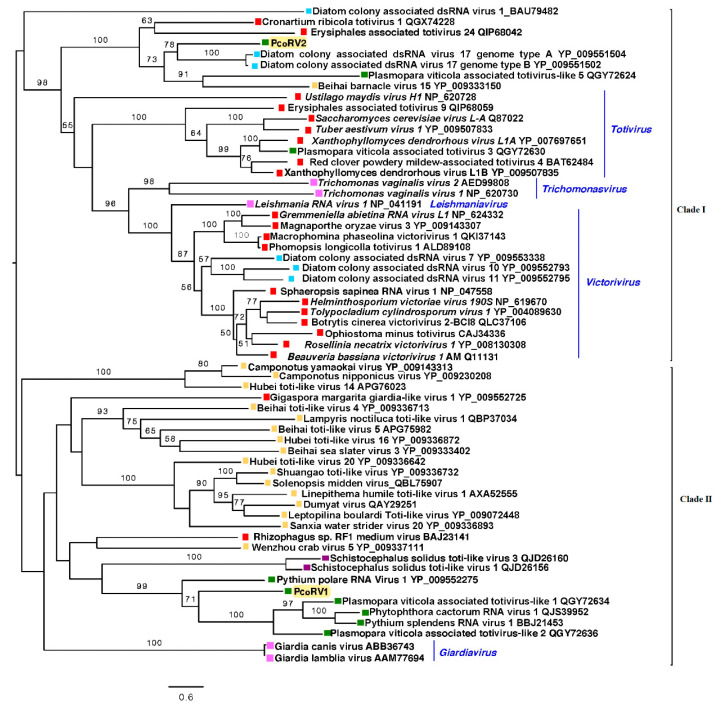
Figure 5.
Phylogenetic analysis (RAxML) based on the predicted totivirus-like RdRp of Phytophthora condilina viruses with other complete classified and unclassified members of the family Totiviridae. Nodes are labelled with bootstrap support values ≥50%. Branch lengths are scaled to the expected underlying number of amino acid substitutions per site. Tree is rooted in the midpoint. Phytopthora condilina RNA virus 1 and 2 are abbreviated (PcoRV1, 2). Family classification and the corresponding pBLAST accession numbers are shown next to the virus names. Colorful squares represent the virus host kingdom or phylum,  Fungi,
Fungi,  Oomycota,
Oomycota,  Arthropoda,
Arthropoda,  Chordata,
Chordata,  Ochrophyta (Heterokonta),
Ochrophyta (Heterokonta),  Excavata. Scale bar = 0.6 expected changes per site per branch.
Excavata. Scale bar = 0.6 expected changes per site per branch.