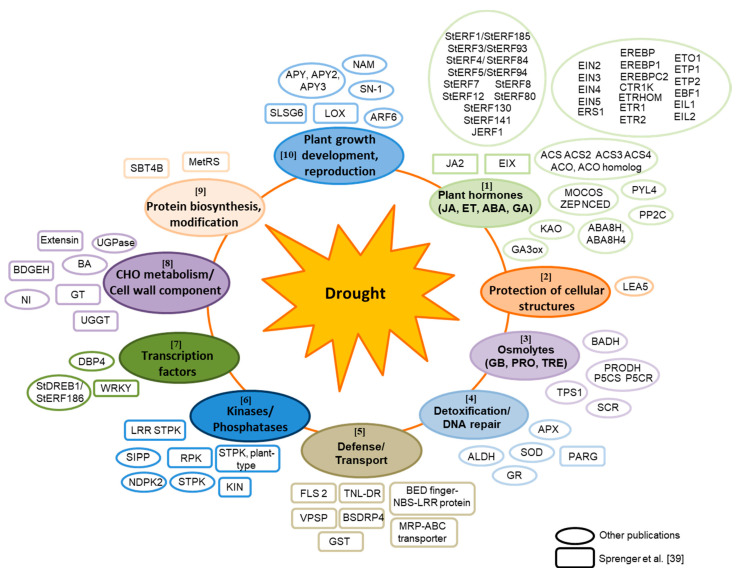
Figure 1.
Overview about processes involved in response to water deficiency showing the selected candidate genes for drought tolerance used in this study for the development of SSR markers. Abbreviations: ABA: Abscisic acid, ABA8H: ABA 8′-hydroxylase, ACO: 1-Aminocyclopropane-1-carboxylic acid oxidase, ACS: 1-Aminocyclopropane-1-carboxylic acid synthase, ALDH: Aldehyde dehydrogenase, APX: Ascorbate peroxidase, APY: Apyrase, ARF: Auxin response factor, BA: β-amylase, BDGEH: β-D-glucan exohydrolase, BADH: Betaine aldehyde dehydrogenase, BSDRP4: Bacterial spot disease resistance protein 4, BEAF: Boundary element associated factor, BED: BEAF and DREF, CHO: Carbohydrate, CTR1K: Constitutive Triple Response1 kinase, DBP: DNA binding protein, P5CS: Delta 1-pyrroline-5-carboxylate synthetase, DREB: Dehydration-Responsive Element-Binding, DREF: DNA replication-related element factor, EBF1: EIN3-binding F-box protein 1, EIN: Ethylene insensitive, EIL: Ethylene-Insensitive3-Like, EREBP: Ethylene-responsive element-binding protein, EREBPC2: Ethylene responsive element binding protein C2, ERF: Ethylene response factor, ERS 1: Ethylene response sensor 1, ET: Ethylene, ETO1: Ethylene-overproduction protein 1, ETP: EIN2 targeting protein, ETR1: Ethylene receptor 1, ETRHOM: Ethylene receptor homolog, EIX: Ethylene-inducing xylanase, FLS 2: Flagellin-sensing 2, GA3ox: Gibberellin 3-oxidase, GA: Gibberellin, GB: Glycine betaine, GR: Glutathione reductase, GST: Glutathione-S-transferase, GT: Glucosyltransferase, JA: Jasmonic acid, JA2: Jasmonic acid 2, JERF1: Jasmonate and Ethylene Response Factor 1, KAO: Ent-kaurenoic acid oxidase, KIN: Kinase, LEA5: Late embryogenesis abundant 5, LOX: Lipoxygenase, LRR: Leucine-rich repeat, MetRS: Methionyl-tRNA synthetase, MOCOS: Molybdenum Cofactor sulfurase, MRP-ABC: Multidrug resistance protein ATP binding cassette, NAM: No apical meristem, NBS: Nucleotide binding site, NI: Neutral invertase, NCED: 9-Cis-epoxycarotenoid dioxygenase, NDPK2: Nucleoside diphosphate protein kinase 2, P5CR: Pyrroline-5-carboxylate reductase, P5CS: Pyrroline-5-carboxylate synthase, PARG: Poly (ADP-ribose) glycohydrolase, PP2C: Protein phosphatase 2C, PRO: Proline, PRODH: Proline dehydrogenase, PYL4: Pyrabactin resistance-like 4, RPK: Receptor protein kinase, SBT4B: subtilase 4B, SCR: SCARECROW, SIPP: Soluble inorganic pyrophosphatase, SLSG6: S-locus specific glycoprotein S6, SN-1: Snaking-1, SOD: Superoxide dismutase, St: Solanum tuberosum, STPK: Serine/threonine protein kinase, TIR: Toll/interleukin-1 receptor-like protein, TNL-DR: TIR-NBS-LRR disease resistance, TPS1: Trehalose-6-phosphate synthase 1, TRE: Trehalose, UGGT: UDP-glucose glycosyltransferase, UGPase: UDP-glucose-1-phosphate uridylyltransferase, VPSP: Vacuolar protein sorting protein, WRKY: WRKY transcription factor, ZEP: Zeaxanthin epoxidase