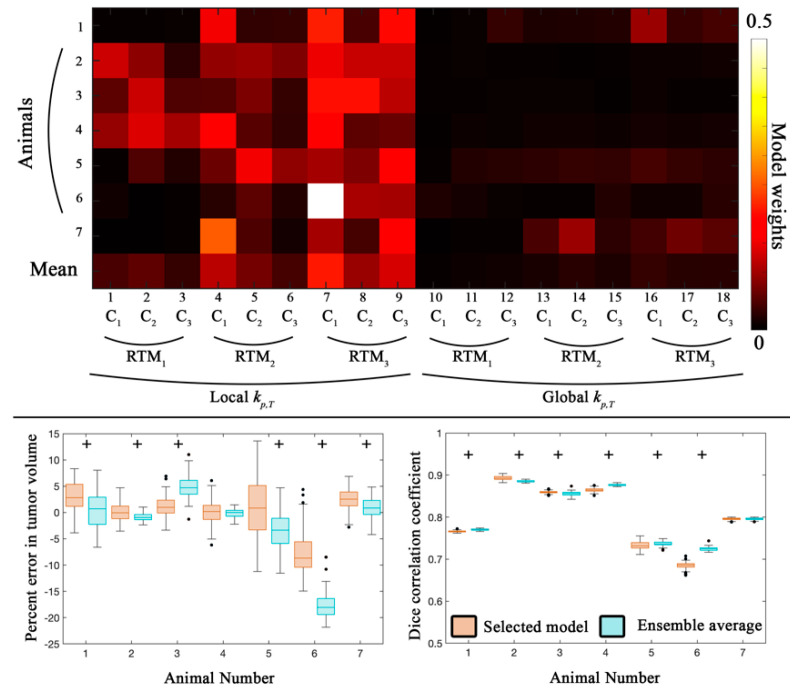
Figure 2.
Global error analysis from the calibration scenario. The top panel shows model weights determined from the Akaike Information Criterion for all seven animals. C1 through C3 correspond to the different approaches to introduce spatial variation in tumor response, while RTM1 through RTM3 correspond to the different RT response models. Models with a local kp,T and RTM3 accounted for 56.9% of the ensemble average model. In the bottom panel, the average percent error in tumor volume and the Dice correlation coefficient are reported for the selected model (orange) and the ensemble averaged model (blue) for all seven animals. The median error for the selected model resulted in less than 9% error for all animals. The median error for the ensemble average was also less than 6% error, with the exception of animal 6 which had greater than 19% error. A strong level of spatial overlap (Dice > 0.68) was observed for all animals. Statistically significant differences (p < 0.05) between model estimates are indicated by the ‘+’ symbol. Model 7 was weighted the highest with an average weight of 0.21.