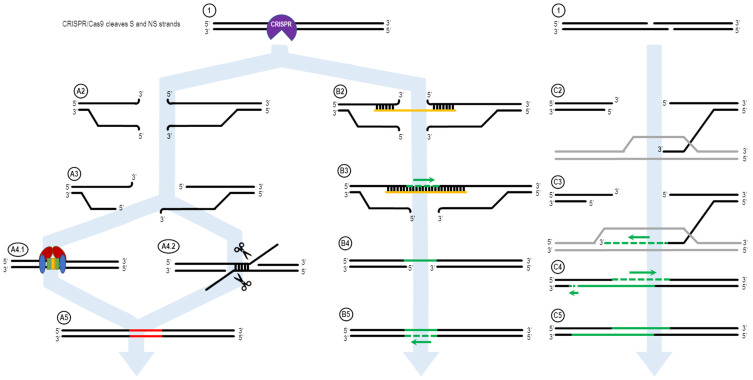
Figure 1.
The ExACT DNA repair pathway, as outlined by Rivera-Torres et al. Upon CRISPR-mediated cleavage or breakage of the DNA (1), several possible pathways may be undertaken in order to facilitate DNA repair. In the absence of an ssODN oligo, or if the ssODN is not in the local vicinity of the broken DNA ends, the non-oligo repair pathway (A2–A5) is used. The CRISPR-Cas complex dissociated from the DNA ends (A2), and resection of the DNA ends may occur (A3). Then, either nonhomologous end joining (NHEJ, A4.1) or microhomology-mediated end joining (MMEJ, A4.2) occurs to repair the broken DNA ends. This results in an indel in the resulting DNA product, as bases are added or deleted via NHEJ and MMEJ. In the presence of an oligo, the ExACT pathway is undertaken (B2–B5). The oligo bridges both sides of the broken DNA ends (B2) and facilitates synthesis-driven repair on one strand (B3). The oligo then dissociated from the repaired DNA strand (B4), so the repaired strand can in turn be used as a repair template for a second synthesis-driven repair event on the other DNA strand (B5). This results in repair containing added, deleted or altered sequence information from the repair template ssODN. In the presence of a homologous gene, synthesis-dependent strand annealing can occur. One 3′-end on either end of the DNA break interrogates the homologous gene and forms a D-loop (C2). The 3′-end is extended via DNA synthesis, expanding the D-loop (C3). After a period of extension, the newly-lengthened 3′-end dissociates from the homologous gene, and binds to its complementary sequence on the other end of the DNA break—bridging the gap (C4). Further DNA synthesis fills in the remaining bases according to their respective complementary sequences (C5).