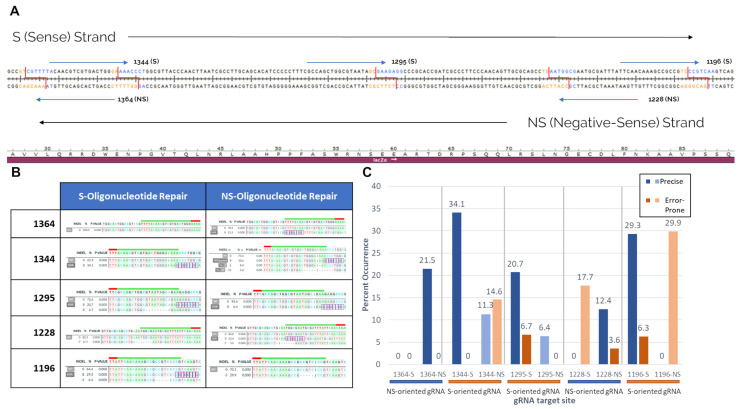
Figure 3.
Secondary non-ExACT products seen in ssODN-containing reactions. (A) Schematic of the LacZ site tested in the in vitro system. Five total sites were analyzed (blue arrows represent gRNA), with the cleavage sites highlighted with red lines. Each cut site was repaired with one of two symmetric ssODNs identical to the sense or negative-sense strand, containing an 8 bp insertion directly at the specific cut site. The bases in yellow and the bases in blue represent the DNA ends resulting from each gRNA cut upstream and downstream, respectively. (B) Resultant repair outcomes for each site, with each ssODN utilized at that site. Deconvolution and alignment of Sanger sequencing products of each in vitro product are displayed, with inserted bases highlighted with purple boxes and deleted bases shown as dashes. Relative percentages of each product are also shown. (C) Quantification of non-HDR indel percentages. For each of the five cleavage sites, the rates of precise (ExACT-driven) and error-prone (MMEJ/NHEJ-driven) products are shown in blue and orange, respectively. For reactions that contained gRNAs and oligos in the same orientation, darker colors are used, while reactions that used gRNAs and oligos of the opposite orientation are displayed with lighter colors.