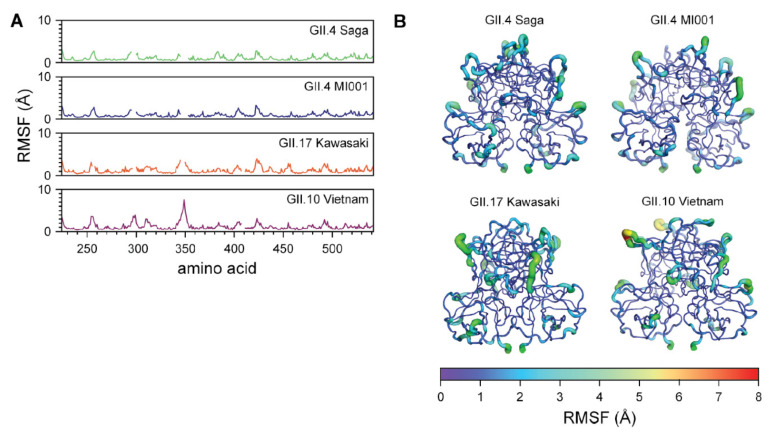
Figure 5.
Difference in fluctuations between the P dimer strains propose increased flexibility in the protein chains in absence of a ligand. (A) RMSF data of GII.4 Saga, GII.4 MI001, GII.17 Kawasaki and GII.10 Vietnam, simulated for a total of 1 µs each, reveal different protein chain dynamics between the strains, with most prominent peak around residue 350. Gaps in the data originate from alignment of the norovirus P dimer sequences. (B) RMSF values of GII.4 Saga, GII.4 MI001, GII.17 Kawasaki and GII.10 Vietnam visualized in the structures, highlighting residues with increased fluctuations during the simulations in absence of ligand.