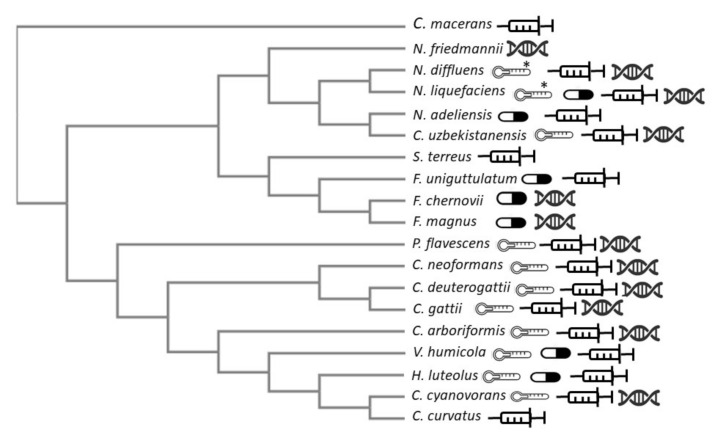
Figure 2.
Neighbor-joining tree without distance corrections obtained using the clustal omega tool (https://www.ebi.ac.uk/). The following Genebank LSU D1/D2 sequences were used: FJ534907 (Cryptococcus deuterogattii CBS 10514T), AF075526 (Cryptococcus gattii CBS 6289T), FJ534909.1 (Cryptococcus neoformans CBS 8710), AB260936 (Cutaneotrichosporon arboriformis CBS 10441T), AF189834 (Cutaneotrichosporon curvatus CBS 570T), JF680899 (Cutaneotrichosporon cyanovorans CBS 11948T), NG_059011.1 (Cystofilobasidium macerans CBS 10757T), AF181530 (Filobasidium chernovii CBS 8679T), AF181851 (Filobasidium magnum CBS 140T), AF075468 (Filobasidium uniguttulatum CBS 1730T), AF075482 (Hannaella luteola/luteolus CBS 943T), AF137603.1 (Naganishia adeliensis CBS 8351T), AF075502 (Naganishia diffluens CBS 160T), AF075478 (Naganishia friedmannii CBS 7160T), AF181515 (Naganishia liquefaciens CBS 968T), AF181508 (Naganishia uzbekistanensis CBS 8683T), AB035042 (Papiliotrema flavescens CBS 942T), AF075479 (Solicoccozyma terreus CBS 1895T), AF189836 (Vanrija humicola CBS 571T). The thermometer graphic represents the species able to grow at 37 °C. * represent poor growth at 37 °C. Pill graphics show species with the highest fluconazole MIC (>16 ug/mL). Syringe graphs show species isolated from normally sterile sites. Double strand graphs show the species identified by molecular-based methods.