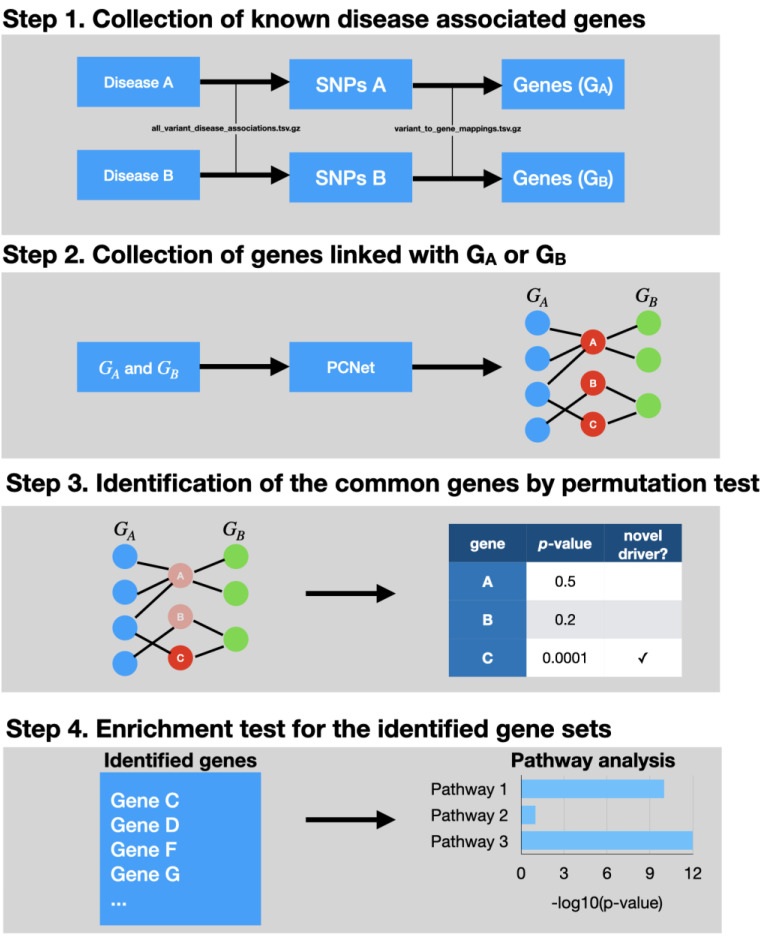
Figure 2.
This figure demonstrates the network-based identification of common driver genes (NICD) workflow, with the specific data sets and programs used construct the algorithm. In the topmost panel, the diseases and genes are identified. These are then placed into PCNet, which is a composite gene network focused on common gene identification for query. In order to remove those genes which maybe randomly associated, a permutation assay with Benjamini–Hoichberg correction is done to ensure that proper candidates demonstrate strength of association. In the bottom-most panel, an enrichment assay is done on the candidate genes which tests their association with canonical pathways.