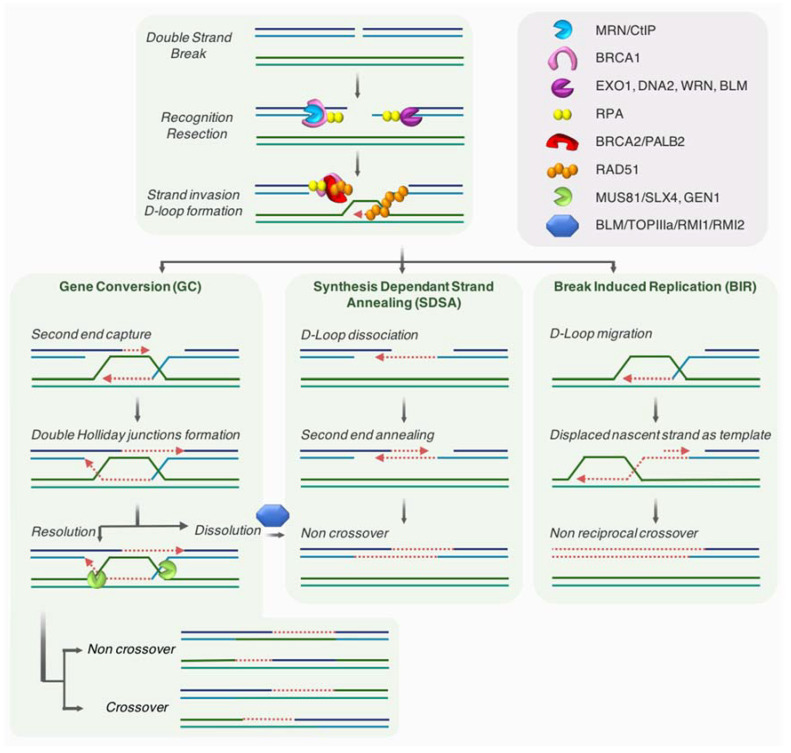
Figure 2.
Homologous-directed repair. The initial resection is mediated by the MRN/CtIP complex then followed by an extensive resection by other helicases and nucleases such as EXO1, DNA2, WRN, or BLM, generating long 3′ ssDNA overhangs. These protruding ssDNA strands are preserved from degradation by the binding of RPA protein. At a later step, both BRCA2/PALB2 promote the replacement of RPA by RAD51, allowing the formation of RAD51 nucleofilament. This RAD51 nucleofilament mediates the homology search, strand invasion and D-Loop formation. Depending on how the intermediates are processed, three main mechanisms have been described as part of the HR pathway: Left panel: In GC, the D-loop extension promotes the annealing of the second end of the break, generating two Holiday junctions (HJ). Dissolution of this two HJ by helicases such as BLM-TOPIIIIa-RMI1-RMI2 will generate non-crossover products, while resolution through cleavage by DNA structure-specific nucleases such as MUS81, SLX4 or GEN1, can mediate both crossover and non-crossover products. Middle panel: In SDSA, the synthesis mediated from the invading 3′ end proceeds until it is sufficient to allow annealing to the 3′ end on the other side of the break, and synthesis of both strand goes on until their ligation with the other end of the break. The D-loop is transitory and no HJ is formed, generating non-crossover products. Right panel: In the BIR pathway, without a second end of the break available for the repair, the synthesis proceeds through the migration of the D-loop and covers long distance. The lagging strand uses as template, the newly synthetized strand emerging from the D-loop. This mechanism can allow the copy of the entire template, resulting in loss of heterozygosity. Blue and green DNA represent sister chromatids and the newly synthetized DNA is highlighted in red.