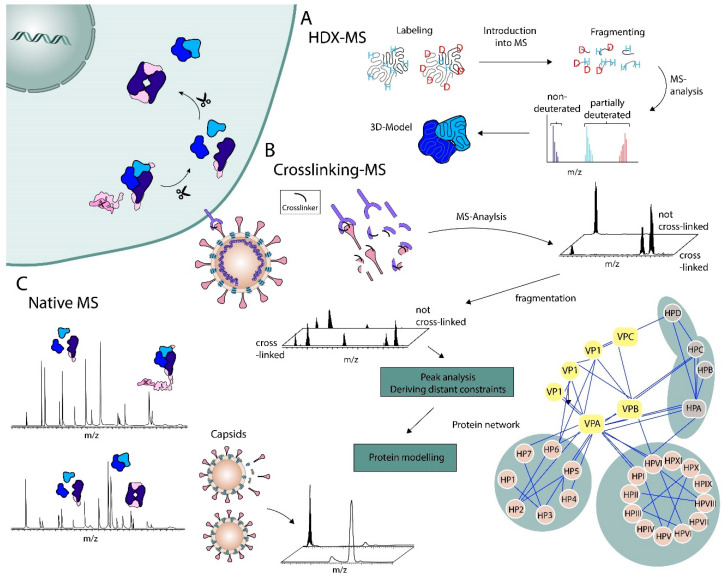
Figure 1.
Structural mass spectrometry (MS) for RNA viruses. All depicted approaches can be conducted as top-down MS whereby exchange of hydrogen to deuterium mass spectrometry (HDX-MS) (A) and crosslinking mass spectrometry (XL-MS) (B) are currently mainly used as bottom-up techniques. For HDX-MS, the workflow starts with labeling of the natively folded protein by exchanging hydrogens to deuterium. The labeled protein is either fragmented or digested to the peptide level. Subsequent MS analyses reveal non-deuterated and partially deuterated peptides leading to constraints for a 3D model. A similar principle is used in XL-MS experiments (B). First, the protein complex is labeled, which can be done in both ways, in vitro and In Vivo, then the sample is fragmented or digested. Distant constraints can be deduced from every successful XL-MS experiment bringing up valuable information for computational modeling and the proposition of a structural model. In Vivo XL-MS offers the identification of a protein interaction network (bottom right) realizing the ability to unravel important virus-host association. (C) Native MS can determine stoichiometries of protein complexes (blue shaded) or measure whole virus capsids.