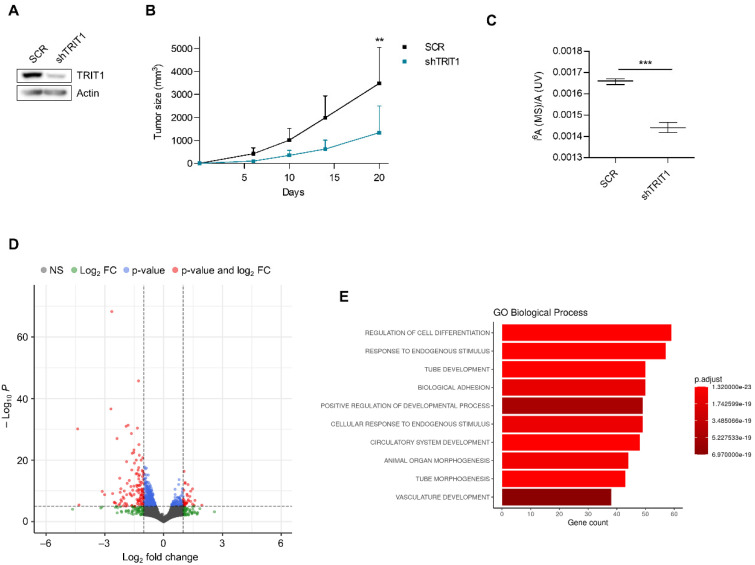
Figure 2.
Effect of TRIT1 depletion on tumor growth and the RNA transcriptome of small-cell lung cancer. (A) Stable downregulation of the TRIT1 gene by short hairpin RNA in the small-cell lung cancer cell line DMS-273 (shTRIT1) determined by western blot analysis. SCR, scramble shRNA. (B) Effect of TRIT1 shRNA-mediated depletion on the growth of subcutaneous tumors in nude mice derived from DMS-273 cells (amplified and overexpressing TRIT1). There was a significant reduction in tumor volume in the TRIT1 shRNA-depleted cells. Data are summarized as the mean and standard deviation (n = 9). Student’s t test, ** p < 0.01. (C) Nucleoside analysis of tRNAs by LC/MS showing that shRNA-mediated depletion of TRIT1 in DMS-273 cells induces the depletion of the i6A-modified nucleoside. Student’s t test, *** p = 0.0002. (D) Volcano plot summarizing the results of the RNA-seq experiment to find mRNAs differentially expressed in TRIT1 shRNA-depleted DMS-273 cells compared with scramble-shRNA DMS-273 cells. (E) Gene ontology (GO) analysis of Biological Process categories in the transcripts downregulated on TRIT1 depletion in DMS-273 cells shows the GO Biological Process category “regulation of cell differentiation” to be the most highly enriched.