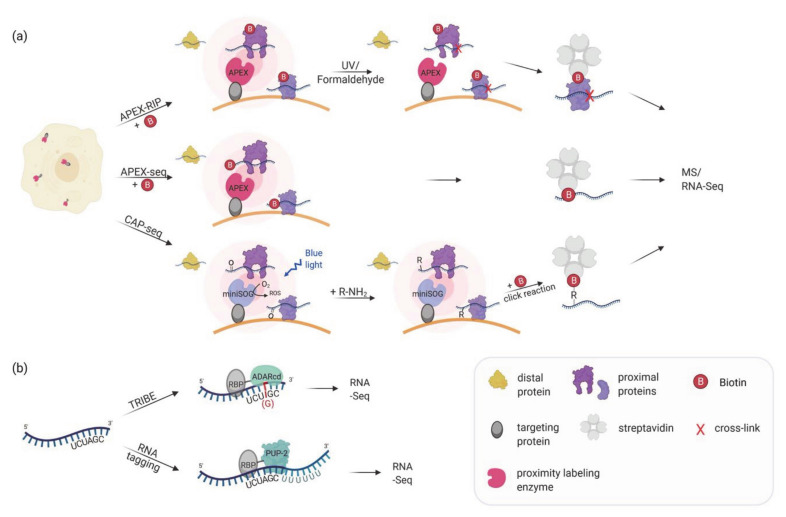
Figure 2.
(a) Proximity labeling strategies for mapping subcellular transcriptomes. APEX or miniSOG are targeted to a specific subcellular location. In APEX-RIP, APEX catalyzes the biotinylation of proximal proteins, and proximal RNAs are crosslinked by either UV light or formaldehyde treatment. In APEX-seq, APEX directly biotinylates RNA. In CAP-seq, miniSOG oxidizes proximal RNA molecules upon blue light illumination. The oxidized RNAs are crosslinked to an alkylamine probe which can be linked to biotin-azide in a click reaction. APEX-RIP, APEX-seq, and CAP-seq make use of streptavidin-purification of the biotinylated proteins/RNAs followed by MS and/or RNA-Seq. (b) In vivo proximity labeling strategies for identifying RNA partners of an RBP. RNA targets of RBPs can be identified by changing the RNA sequence upon the interaction. In TRIBE the RBP of interest is fused to the catalytic domain of ADAR which mediates adenosine to inosine editing (in red) of the interacting RNA(s). The inosine is read as a guanosine when analyzed by RNA-Seq allowing the identification of editing events as A-to-G transition. In RNA tagging the RBP of interest is fused to a poly(U) polymerase (PUP-2) that attaches a poly-uracil chain at the 3′end of interacting RNAs. The uracil tail is then used to identify targets during RNA sequencing.