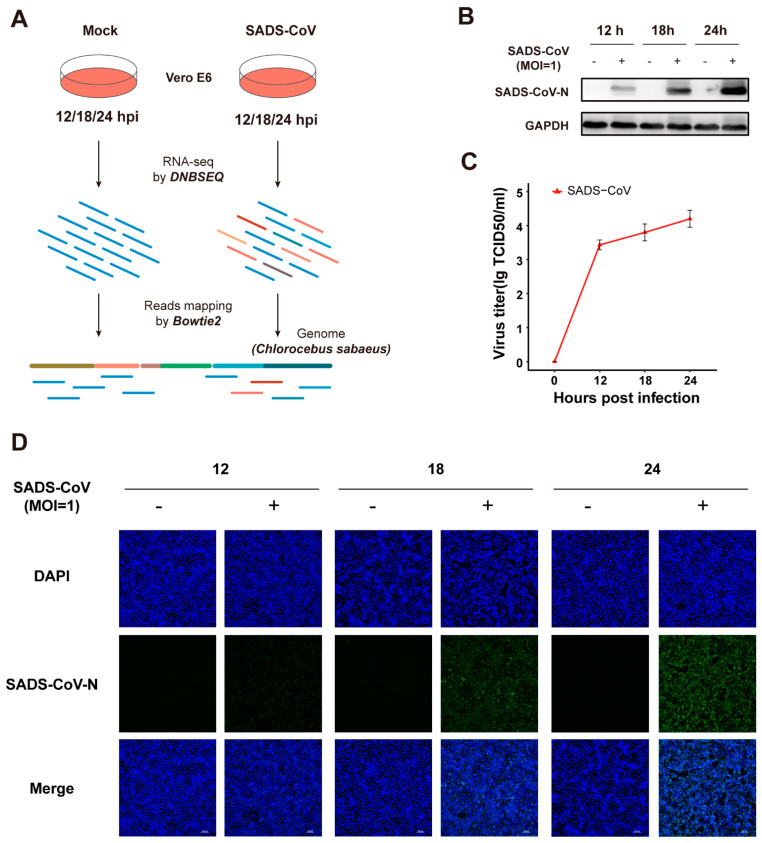
Figure 1.
Workflow and confirmation of productive swine acute diarrhea syndrome coronavirus (SADS-CoV) infection in cultured Vero E6 cells. (A) Workflow. Vero E6 cells were mock-infected or infected with SADS-CoV strain GDS04 (MOI = 1), followed by sample collection at 12, 18, 24 hpi. Samples from each group were prepared in triplicate. Total RNA was extracted and BGI DNBSEQ RNA-sequencing was performed. (B) Western blot confirmation via viral N-protein in SADS-CoV-infected Vero E6 cells. Cells were fixed at 12, 18, or 24 hpi, and N protein was detected by Western blot; GAPDH was conducted as an internal control gene. (C) The virus titer of SADS-CoV in Vero E6 cells was measured by TCID50. (D) Immunofluorescence staining of the viral N-protein in SADS-CoV-infected Vero E6 cells. Cells were fixed at 12, 18, or 24 hpi, and N protein (green) was detected by indirect immunofluorescence assay. Nuclei (blue) were shown by 4′,6-diamidino-2-phenylindole (DAPI) staining. The images of cells were acquired by a fluorescence microscope (Nikon) at a 40× magnification.