Abstract
Purpose
Hepatocellular carcinoma (HCC) has the fourth highest rate of mortality among the different types of cancer worldwide. This study aimed to investigate the functions of microRNA-145-5p and AFR6 on migration, invasion and metastasis in HCC.
Methods
A total of 150 pairs of tumor and their matched adjacent nontumor liver tissues were collected from HCC patients. Expressions of microRNA-145-5p and AFR6 were measured by real-time PCR in HCC tissues and in HCC cell lines. The correlations between microRNA-145-5p and HCC prognosis were investigated. The proliferation, migration, invasion, cell cycle progression, and apoptosis of HCCLM3 cells were evaluated with CCK8, wound healing, transwell, and flow cytometric experiments.
Results
The expression of miR-145-5p was confirmed to be downregulated not only in HCC tissues but also in several HCC cell lines compared with normal controls. A low expression level of miR-145-5p was notably associated with poor prognosis in patients with HCC and certain characteristics of metastatic tumors. In vitro, miR-145-5p negatively regulated cell proliferation, migration, and invasion and induced apoptosis in HCCLM3 cells. Subsequent experiments further verified that ARF6 is a novel target of miR-145-5p and is significantly overexpressed in HCC tissues. Overexpression of ARF6 circumvented the effects of miR-145-5p in HCCLM3 cells.
Conclusion
miR-145-5p may play a pivotal role in HCC metastasis via regulating ARF6, and these findings may both provide further insights into the key factors of HCC metastasis and prove to be useful in the development of novel treatment options for HCC.
Keywords: microRNA-145-5p, ADP-ribosylation factor 6, hepatocellular carcinoma, proliferation, invasion, metastasis
Introduction
Among malignancies worldwide, liver cancer has been reported to be the sixth most frequently diagnosed cancer and has the fourth highest mortality rate, causing ~780,000 deaths each year.1 Despite comprehensive approaches through multiple modalities, such as surgical excision, interventional therapy, and targeted therapy, liver cancer treatment still produces unsatisfactory results. Hepatocellular carcinoma (HCC) metastasis provides one of the major reasons why patients miss the opportunity for optimal treatment and ultimately succumb to mortality.2 Therefore, the exploration of the internal principles underlying the occurrence, development and metastasis of HCC and the research for new prognostic markers and prevention targets have important clinical significance for improving the cure rate of HCC. HCC invasion and metastasis are complex processes involving multiple factors and genes, although the specific mechanisms have yet to be fully elucidated.3
microRNAs (miRNAs/miRs) are highly conserved small non-coding RNAs of ~17-25 nucleotides in length.4 They can bind to the 3ʹ untranslated regions (UTRs) of target genes, thereby negatively regulating mRNA expression.5 Abnormal expression of miRNAs is involved in regulating gene expression, cell differentiation, cell proliferation, and the invasion and metastasis of malignant tumors through mRNA degradation and translational suppression.6 Moreover, miR-145-5p has been shown to exert antitumor effects in several different types of cancer.7 When comparing miRNA dysregulation in different molecular subtypes, it was found that miR-145-5p is among the most notably suppressed miRNAs in HCC.8 Recently, some studies have demonstrated that miR-145 inhibits cell migration and invasion, possibly by targeting Golgi membrane protein 1 (GOLM1).9 Although the role of miRNAs in HCC has attracted extensive research attention, the underlying mechanisms and significance of miRNA-145-5p in HCC remain unclear.
ADP-ribosylation factor 6 (ARF6) is a GTP-binding protein in the Ras superfamily of small GTPases and is a member of the ARF family.10 It participates in the regulation of cell division, microvesicle formation and transportation, and cell adhesion and is closely associated with tumor cell growth, migration and metastasis.11 ARF6 is abnormally expressed at a high level in numerous types of malignant tumor cells and tissues, such as those of pancreatic cancer and laryngeal squamous cell carcinoma, and is involved in regulating the development of multiple tumor types.12,13 It has been reported that intracellular circulation driven by ARF6 may control the malignant phenotype of HCC, although the expression level and underlying specific mechanism of ARF6 in HCC remain unclear.14 The epidermal growth factor receptor (EGFR)/GEP100/ARF6 pathway has been demonstrated to have a significant role in the metastasis of numerous types of tumor and is associated with prognosis in diseases, such as breast cancer, squamous cell carcinoma, and glioblastoma.15–17 Therefore, the present study aimed to explore the role of ARF6 and its regulatory mechanism in HCC and to facilitate the identification of effective treatments for the prevention of HCC invasion and metastasis.
In the present study, it was first demonstrated that miRNA-145-5p is downregulated in HCC tissues and cells. Dysregulation of miR-145-5p was found to be markedly correlated with poor prognosis in patients with HCC. Furthermore, it was shown that miRNA-145-5p inhibits HCC cell migration and invasion in HCC through targeting ARF6.
Materials and Methods
Surgical Specimens and Clinicopathological Data
Between April 2011 and October 2013, primary cancer tissues were pathologically confirmed as being HCC (n=150), and their matched adjacent nontumor liver tissues (ANLTs) were obtained from the First Affiliated Hospital, Zhejiang University School of Medicine. It was ensured that the distance between the ANLTs and corresponding tumor edges was greater than 2 cm. The clinical characteristics of the HCC patients are described in Table 1. After surgery, HCC patients were followed up for 1–70 months. Disease-free survival rates of the patients were determined according to the operation date and the date of recurrent disease diagnosis or death. Survival rates were determined from the operation date to the date of death or the end of follow-up. Cellular differentiation stages were determined by Edmondson grading. Normal liver samples were collected from the normal parts of resected specimens from patients with hepatic hemangiomas. All tissues were stored at −80 °C prior to use, and the tissues were acquired with written informed consent from the patients. The outline of the research project was approved by the Clinical Research Ethics Committee of the First Affiliated Hospital, Zhejiang University School of Medicine, in accordance with the Declaration of Helsinki.
Table 1.
Correlation of miR-145-5p Expression with Clinicopathological Characteristics in Hepatocellular Carcinoma
| Variables | Cases (n=150) | miR-145-5p Expression | P value | |
|---|---|---|---|---|
| High (n = 75) | Low (n = 75) | |||
| >0.386 | <0.386 | |||
| Age (years) | 0.407 | |||
| ≤55 | 62 | 34 | 28 | |
| >55 | 88 | 41 | 47 | |
| Sex | 0.449 | |||
| Male | 113 | 59 | 54 | |
| Female | 37 | 16 | 21 | |
| Tumor size (cm) | 0.019a | |||
| ≤5 | 91 | 53 | 38 | |
| >5 | 59 | 22 | 37 | |
| Cellular differentiation | 0.022a | |||
| I–II | 113 | 63 | 50 | |
| ш–IV | 37 | 12 | 25 | |
| Tumor encapsulation | 0.022a | |||
| Present | 48 | 31 | 17 | |
| Absent | 102 | 44 | 58 | |
| HBV infection | 0.464 | |||
| Present | 109 | 52 | 57 | |
| Absent | 41 | 23 | 18 | |
| Portal vein tumor embolus | 0.018a | |||
| Present | 42 | 14 | 28 | |
| Absent | 108 | 61 | 47 | |
| TNM tumor stage | 0.013a | |||
| I–II | 103 | 59 | 44 | |
| ш–IV | 47 | 16 | 31 | |
Abbreviations: TNM, tumor-node-metastasis; HBV, hepatitis B virus; a Statistically significant.
Cell Culture and Transfection
Our study used several HCC cell lines. Huh7, MHCC97L, MHCC97H, and HCCLM3 cells were purchased from Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, China). Hep3B cells and the normal liver cell line THLE-2 were purchased from the ATCC (Manassas, VA, USA). All cells were cultured in DMEM (Gibco; Thermo Fisher, Carlsbad, CA, USA) according to the manufacturer’s recommendations. The hsa-miR-145-5p mimics (miR10000437-1-5), hsa-miR-145-5p inhibitor (miR20000437-1-5) and negative control vector were obtained from RiboBio (Guangzhou, China). The sequences of the miRNAs were as follows: 5ʹ-GUCCAGUUUUCCCAGGAAUCCCUU-3ʹ for the miR-145-5p mimics; 5ʹ-ATCGGAATTCGGACAGCCTTCTTC-3ʹ for the miR-145-5p inhibitor; 5ʹ-TTCTCCGAACGTGTCACGT-3ʹ for the mimics negative control vector; and 5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ for the inhibitor negative control vector. ARF6 lentiviral activation particles (sc-400799-LAC), which could specifically and efficiently upregulate gene expression when introduced into cells via lentiviral transduction, and control lentiviral activation particles (sc-437282) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). HCCLM3 cells were transfected with the miR-145-5p mimics or negative control vector or were cotransfected with the miR-145-5p mimics and either ARF6 lentiviral activation particles or control lentiviral activation particles. THLE-2 cells were transfected with the negative control vector or hsa-miR-145-5p inhibitor. Cells were transfected 24 h before experiments.
Cell Counting Kit-8 (CCK-8) Assay
Two groups of HCCLM3 cells were digested with digestive enzymes and reseeded in 96-well plates. Then, ~400,000 cells were added to each well and the cells were cultured in an incubator for 24, 48 or 72 h. Subsequently, CCK-8 reagent was added to the cells followed by mixing, and the cells were then incubated for 1 h. A microplate reader (Dojindo Molecular Technologies, Kumamoto, Japan) was applied to estimate the absorbance at 450 nm in order to generate a growth curve.
Flow Cytometric Analysis of Apoptosis
HCCLM3 cells were harvested after 48 h, and apoptosis rates were detected using an Annexin-V/FITC kit (BD Biosciences, San Jose, CA, USA). Briefly, HCCLM3 cells were washed with pre-chilled PBS, followed by the addition of 1 mL 1X Annexin V-FITC binding buffer. The mixture was centrifuged, and then resuspended by the addition of 200 μL of binding buffer. The HCCLM3 cells were subsequently treated with 10 μL annexin V-FITC and 5 μL PI, and incubated for 30 min. Finally, the apoptosis rates in each group were determined using a FACSCalibur flow cytometer (BD Biosciences).
Cell Cycle Analysis
HCCLM3 cells of the three groups were collected at a density of ~1,000,000 cells/mL, seeded into 6-well plates. After washing with PBS, the cells were fixed in 75% ethanol at 4 °C overnight. Then, HCCLM3 cells were washed with PBS twice, followed by the incubation with propidium iodide working solution. The cells were subjected to cell cycle analysis using FACSCalibur instrument (BD Biosciences).
Transwell Chamber Assay
Matrigel™ (BD Biosciences) was added to the upper chambers of Transwell inserts (Corning Inc, Beijing, China). After 24 h of transfection, HCCLM3 cells were trypsinized and resuspended in serum-free medium. In total, ~40,000 cells were added to the upper chambers, and 500 μL DMEM was added to the lower chambers. After a wash with PBS, the noninvaded cells in the upper chambers were gently wiped with a wet cotton swab, and the remaining cells were fixed with 4% paraformaldehyde for 20 min. After washing 3 times with PBS, the cells were stained with 0.1% methyl violet for 25 min. After washing, the stained cells were counted under a microscope in 5 randomly selected fields.
Wound Healing Assay
Transfected HCCLM3 cells or THLE-2 cells were evenly spread on to a 6-well plate (~500,000 cells per well). When the cells were 90% confluent, the cell layer was uniformly scratched with a 200 μL pipette tip. Subsequently, the cells were observed under a microscope and imaged after the floating cells were removed. After 24 h, the cells were observed again, and the migration area was measured in 5 random fields.
Database Analysis
The online target gene prediction databases TargetScan 7.2 (http://www.targetscan.org/vert_72/) and miRanda (http://www.microrna.org/microrna/home.do) were used to predict the miR-145-5p target genes and subsequently determined the intersection of the results of the two predictions. In addition, target gene databases with experimental verification support, including miRTarBase (http://mirtarbase.mbc.nctu.edu.tw) and miRwalk (http://129.206.7.150/), were jointly used to identify possible targets of miR-145-5p. The results were combined as the final miR-145-5p target gene prediction result.
Dual-Luciferase Reporter Assay
A luciferase reporter assay with the ARF6 3ʹ-UTR was used to verify that ARF6 is the direct target of miR-145-5p. The negative control vector or miR-145-5p mimics was co-transfected with the constructed wild-type (WT)-ARF6-3ʹ-UTR (WT seed sequence: AACUGGA) or mutated (MT)-ARF6-3ʹ-UTR (MT seed sequence:UUGACCU) psiCHECK2 reporter vector (Promega, Madison, WI, USA) into HCCLM3 cells using Lipofectamine 2000®, according to the manufacturer’s instructions. Renilla and firefly luciferase activities were assessed using a dual-luciferase reporter assay system (Promega).
RNA Extraction and Reverse Transcription-Quantitative PCR (RT-qPCR) Analyses
Total RNA was extracted from HCC frozen tissues and HCC cell lines using the TRIzol® method (Invitrogen; Thermo Fisher Scientific, CA, USA) and stored in a freezer at −80 °C. Subsequently, the RNA was reverse-transcribed to complementary DNA (cDNA) with a miRNA qPCR detection kit (GeneCopoeia, USA) according to the manufacturer’s instructions. The primer sequences used for RT-qPCR were as follows: 5ʹ-GTCCAGTTTTCCCAGGAATCCCT-3ʹ (forward) and 5ʹ-TCCAGTCCTATTGAATGTGGGA-3ʹ (reverse) for miR-145-5p, 5ʹ-ATGGGGAAGGTGCTATCCAAAATC-3ʹ (forward) and 5ʹ-GCAGTCCACTACGAAGATGAGACC-3ʹ (reverse) for ARF6, and 5ʹ-CTCGCTTCGGCAGCACA-3ʹ (forward) and 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ (reverse) for U6. ARF6 data were normalized against GAPDH, which was detected using the following primers: 5ʹ-GCACCACCAACTGCTTAGCA-3ʹ (forward) and 5ʹ-TCTTCTGGGTGGCAGTGATG-3ʹ (reverse). U6 was used as the endogenous control for miR-145-5p.
Immunohistochemical (IHC) Staining of ARF6
Formalin-fixed and paraffin-embedded HCC tissues were cut into 4-μm-thick sections and were subsequently subjected to antigen retrieval and quenching of endogenous peroxidase activity. Antibody against ARF6 (1:300; Santa Cruz Biotechnology) was used as the primary antibody; the slides were incubated at 4°C overnight and were subsequently incubated with a secondary antibody (1:500; MultiSciences, Hangzhou, China). Finally, DAB solution was applied for signal generation, and the slides were stained again with hematoxylin and observed under an optical microscope. Immunohistochemical staining was assessed by two pathologists who were blinded to the patients’ clinical data to independently determine whether the patients were positive for ARF6 expression.
Statistical Analysis
Data are expressed as the mean ± SEM values and were analyzed using SPSS 18.0 software (IBM Corp.). Experiments were performed ≥3 times. The χ2 test was used to explore the association between the two variables. One-way ANOVA was used to compare the three experimental groups, with the Student-Newman-Keuls test and Tukey HSD for pairwise inter-comparison of the subgroups. The overall survival and disease-free survival rates of the patients were measured using the Kaplan–Meier method, and the survival rate between the groups was compared by Log rank test. Correlation of miR-145-5p levels with ARF6 mRNA expressions was examined by the Pearson correlation. P <0.05 was considered to indicate a statistically significant difference.
Results
miR-145-5p is Downregulated in HCC Tissues and Cells
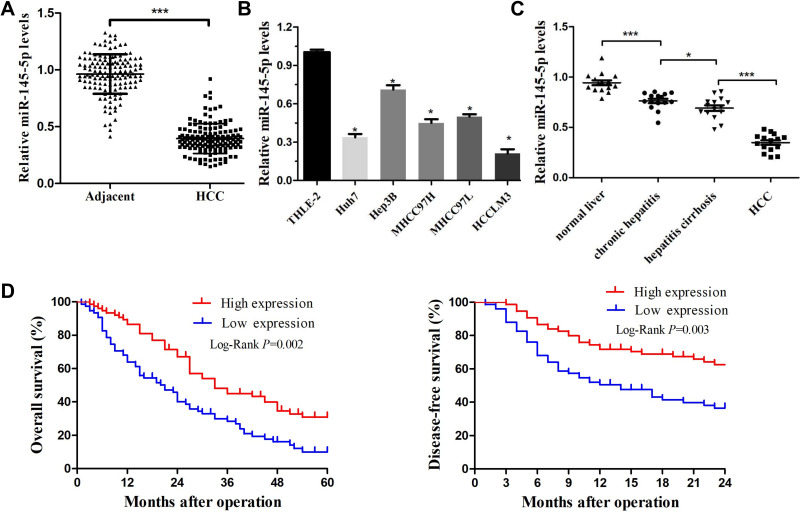
In previous studies, miRNA-145-5p was often found to be negatively regulated in HCC tissues. To further verify this observation, the expression level of miRNA-145-5p (hereafter abbreviated miR-145) was first measured in 150 pairs of HCC tissues and matched ANLTs by RT-qPCR. Notable downregulation of miR-145 was observed in HCC tissues compared with the matched ANLTs (Figure 1A). To explore whether miR-145 is dysregulated in HCC cells, the expression levels of miR-145 in HCC cell lines (Huh7, MHCC97L, MHCC97H, HCCLM3, and Hep3B) and normal liver THLE-2 cells were quantified. It was confirmed that miR-145 expression was also reduced in HCC cell lines compared with THLE-2 cells (Figure 1B). Therefore, these experiments demonstrated the frequent downregulation of miR-145 in HCC. Subsequently, whether miR-145 is gradually downregulated during the development of HCC was further examined. To this end, the samples were divided into the following 4 groups: The normal liver group, the nontumor chronic hepatitis group, the hepatitis cirrhosis group, and the HCC group. The results demonstrated that the miR-145 expression decreased notably as HCC progressed. miR-145 expression was highest in normal liver tissues, gradually decreased from nontumor chronic hepatitis tissues to hepatitis cirrhosis tissues, and was lowest in HCC tissues (Figure 1C). These findings revealed that miR-145 may participate in the development of HCC.
Figure 1.
miR-145-5p was downregulated in HCC and was associated with poor prognosis. (A) Expression of miR-145-5p was measured by real-time PCR in HCC tissues and their matched ANLTs. ***P<0.001 compared with ANLTs. (B) Expression of miR-145-5p in HCC cell lines (Huh7, MHCC97L, MHCC97H, HCCLM3, and Hep3B) and a normal liver cell line (THLE-2). *P<0.05 compared with the THLE-2 cell line. (C) RT-qPCR was used to detect miR-145-5p expression in the normal liver group, nontumor chronic hepatitis group, hepatitis cirrhosis group, and HCC group (n=15 per group). Blood tests were used to diagnose whether the patient has HBV infection. Ultrasound, CT scan and liver biopsy were used to assess whether the patients had cirrhosis. Patients without hepatitis and cirrhosis were included in the normal liver group. Hepatitis patients without cirrhosis and HCC were included in the nontumor chronic hepatitis group. Hepatitis cirrhosis patients without HCC were included in the hepatitis cirrhosis group. *P<0.05 and ***P<0.001 compared among groups. (D) After 5 years of follow-up, the 2-year disease-free survival rate and 5-year overall survival rate of HCC patients were calculated by the Kaplan–Meier method. The red line represents the high miR-145 high expression group. The blue line represents the low miR-145 expression group. The data are presented as the mean ± standard deviation values.
Abbreviation: HCC, hepatocellular carcinoma.
Downregulation of miR-145 is Associated with a Poor Prognosis
To reveal the clinicopathological characteristics of miR-145 in HCC, patients with HCC were divided into two groups according to the median relative expression level of miR-145-5p (0.386). HCC tissues with a miR-145-5p level greater than 0.386 were assigned to the high expression group. Those with a miR-145-5p level below 0.386 were assigned to the low expression group. The results suggested that low expression of miR-145 was markedly associated with invasive pathological features of HCC, including larger tumor size, poorer cellular differentiation, lack of tumor encapsulation, portal vein tumor embolus, higher tumor stage, and cancer death (Table 1). Because portal vein tumor embolus and advanced tumor stage are associated with HCC metastasis, downregulation of miR-145 may be involved in the metastatic growth of HCC. In addition, to reveal the relationship between miR-145 downregulation and patient prognosis, the correlation between low miR-145 expression and patient survival was also investigated. After 5 years of follow-up, it was found that the 2-year disease-free survival rates and 5-year overall survival rates of patients in the low miR-145 expression group were notably lower than those of patients in the high expression group (Figure 1D). Collectively, the above data revealed that loss of miR-145 expression may promote HCC metastasis and this was correlated with the poor prognosis of patients with HCC.
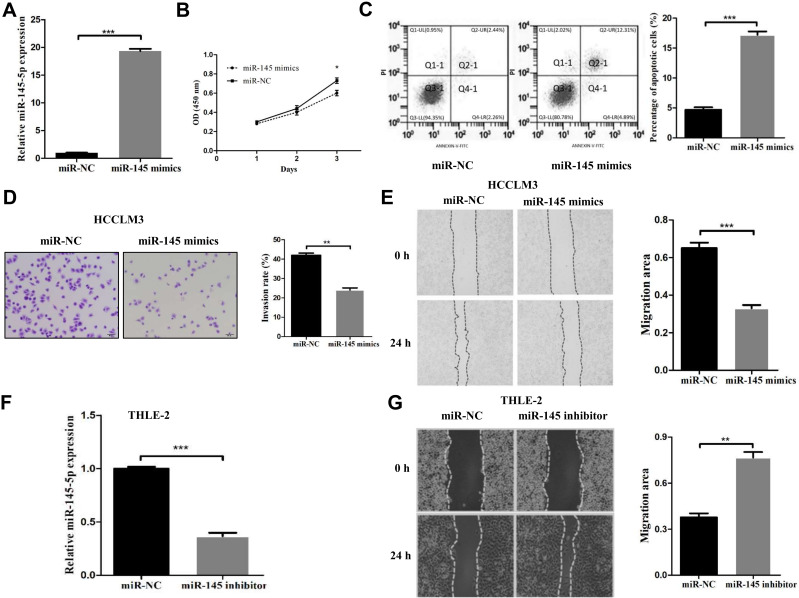
miR-145 Negatively Regulates HCCLM3 Cell Proliferation, Migration, and Invasion and Induces Apoptosis
Since it was observed that deletion of miR-145 was closely correlated with the metastasis and prognosis of HCC, and tumor cell proliferation, apoptosis and invasion have important roles in tumor growth and metastasis, it was possible to hypothesize that restoration of miR-145 levels may inhibit HCC cell proliferation, migration and invasion. Owing to the most marked downregulation of miR-145 in HCCLM3 cells, HCCLM3 cells that could stably express miR-145 were first established. Compared with that in the negative control group, miR-145 expression was confirmed to notably increase in the miR-145 mimic transfection group (Figure 2A). Using CCK-8 assay, it was observed that miR-145 re-expression notably inhibited HCCLM3 cell proliferation in vitro (Figure 2B). Flow cytometric analysis was subsequently used to reveal the effect of miR-145 re-expression on the apoptosis rates. The data indicated that the apoptosis rate of HCCLM3 cells was increased markedly with increasing miR-145 expression, which demonstrated that miR-145 induced apoptosis in HCC cells (Figure 2C). In the Transwell cell invasion assay, overexpression of miR-145 reduced the number of cells that invaded into the lower chamber (Figure 2D). Similarly, in the miR-145 high expression group, the ability of cells to migrate was notably impeded compared with that in the low expression group (Figure 2E). To further detect the effects of miR-145 on cell migratory and invasive abilities, THLE-2 cells expressing relatively high levels of endogenous miR-145 were transfected with miR-145 inhibitors. Downregulation of miR-145 was verified by RT-qPCR (Figure 2F). Interestingly, inhibition of miR-145 markedly promoted the migration of THLE-2 cells (Figure 2G). Our results described above demonstrated that miR-145 suppressed HCC metastasis by negatively regulating cell proliferation, migration, and invasion and inducing apoptosis.
Figure 2.
miR-145 inhibited proliferation, migration, and invasion and induced apoptosis in HCC cell lines. (A) The miR-145-5p mimics and negative control were efficiently transfected into HCCLM3 cells. (B) Cell proliferation rates were measured in two groups of HCCLM3 cells by a CCK-8 assay after 24 h, 48 h and 72 h. (C) Flow cytometric analysis was used to determine the apoptosis rates of HCCLM3 cells. (D) The invasive ability of transfected HCCLM3 cells was evaluated by a Transwell assay (magnification, 200×). (E) Migration rates of HCCLM3 cells were determined by a wound healing assay. (F) The expression of miR-145-5p was measured in THLE-2 cells transfected with the miR-145-5p inhibitor or negative control. (G) Migration rates of THLE-2 cells were confirmed by a wound healing assay. *P<0.05, **P<0.01 and ***P<0.001 compared with the NC group. The data are presented as the mean ± standard deviation values.
Abbreviation: NC, negative control.
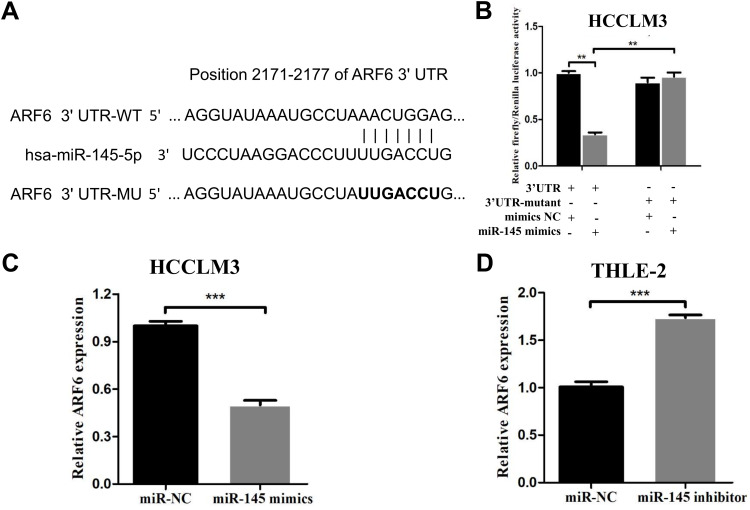
ARF6 is a Direct Target of miR-145 in HCCLM3 Cells
Previous studies have revealed that miR-145 functions as a tumor suppressor gene in HCC. Accordingly, the underlying targets of miR-145 that are associated with tumor metastasis were sought after. miR-145 downstream gene targets were searched for using four different miRNA targeting prediction tools: TargetScan, miRanda, miRTarBase and miRwalk. A potential candidate target gene, ARF6, was subsequently selected, which has a predicted consequential pairing with miR-145 at nucleotide positions 2171–2177 (Figure 3A). To further explore whether the antimetastatic properties of miR-145 in HCC are associated with ARF6, a dual-luciferase reporter assay in HCCLM3 cells was first performed. The luciferase activity in HCCLM3 cells with wild-type ARF6 following transfection with the miR-145 mimics was markedly reduced compared with that in mimic NC-transfected cells. However, in HCCLM3 cells with mutated ARF6, the decrease in luciferase activity was significantly reversed (Figure 3B). Subsequently, whether miR-145 could regulate ARF6 expression at the cellular level is investigated. After transfection with miR-145 mimics, ARF6 levels in HCCLM3 cells were decreased by ~50% (Figure 3C). Inhibition of miR-145 in normal liver THLE-2 cells led to a marked increase in ARF6 expression (Figure 3D). Accordingly, ARF6 was indicated to be a novel target of miR-145 in HCCLM3 cells.
Figure 3.
ARF6 was proven to be a novel target of miR-145-5p in HCCLM3 cells. (A) ARF6 had a predicted consequential pairing with miR-145-5p at nucleotide positions 2171–2177. psiCHECK2 dual-luciferase reporter vectors containing WT or MT ARF6 3ʹ-UTR were constructed. (B) Renilla luciferase activity was normalized to firefly luciferase activity in HCCLM3 cells expressing WT-ARF6 or MT-ARF6. (C) After transfection with miR-145 mimics, the ARF6 levels were significantly decreased in HCCLM3 cells. (D) RT-qPCR was performed to measure the expression level of ARF6 in THLE-2 cells. **P<0.01 compared with the 3ʹ-UTR+miR-145 mimics group and ***P<0.001 compared with the NC group. The data are presented as the mean ± standard deviation values.
Abbreviations: ARF6, ADP-ribosylation factor 6; NC, negative control; UTR, 3ʹ untranslated regions; WT, wild-type; MT, mutated.
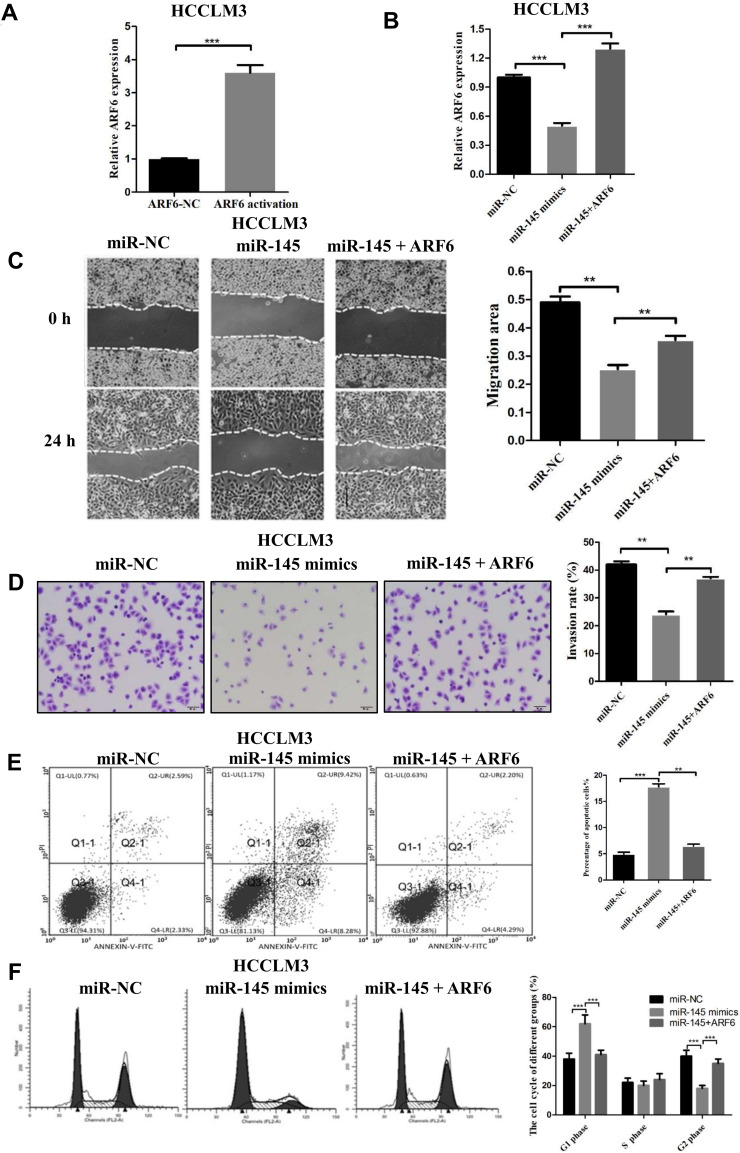
Overexpression of ARF6 Circumvents the Effects of miR-145 in HCCLM3 Cells
The above findings indicated that miR-145 inhibits metastasis by negatively regulating HCC cell migration and invasion and that ARF6 is directly targeted by miR-145. Therefore, it was hypothesized that ARF6 may participate in miR-145-mediated antimetastatic effects. To confirm that the transfections are successful with lentiviral activation particles, HCCLM3 cells were first transfected with ARF6 lentiviral activation particles or control lentiviral activation particles. The mRNA level of ARF6 increased after transfection of ARF6 lentiviral activation particles (Figure 4A). To verify that miR-145 directly targets ARF6 to perform its function, miR-145-transfected HCCLM3 cells were infected with ARF6 lentiviral activation particles or control lentiviral activation particles. ARF6 mRNA levels were significantly increased by transfection of ARF6 lentiviral activation particles (Figure 4B). In the wound healing assays, restoration of ARF6 expression partially promoted the migration of HCCLM3 cells overexpressing miR-145 (Figure 4C). In the Transwell assays, increased expression of ARF6 enhanced the invasive ability of miR-145-overexpressing HCCLM3 cells, which was suppressed by miR-145 (Figure 4D). Moreover, apoptosis induced by miR-145 was reversed by ARF6 (Figure 4E). In the cell division cycle analysis, the miR-145 re-expression increased the percentage of the HCCLM3 cells at the G1 phase and decreased the ratio of cells at the G2 phase. But the overexpression of ARF6 circumvented these effects (Figure 4F). Collectively, these results suggested that miR-145 inhibited invasion and metastasis by directly targeting ARF6.
Figure 4.
Overexpression of ARF6 counteracted the effects of miR-145-5p in HCCLM3 cells. (A) The mRNA level of ARF6 increased after transfection of ARF6 lentiviral activation particles. ***P<0.001 compared with the control lentiviral activation particles. (B) ARF6 levels were measured by RT-qPCR in HCCLM3 cells. (C) Migration rates of HCCLM3 cells were confirmed by a wound healing assay. (D) The invasive ability of transfected HCCLM3 cells was measured by Transwell assay (magnification, 200×). (E) Apoptosis induced by miR-145 was reversed by ARF6. (F) The effects of miR-145 and ARF6 on the cell division cycle were measured by cell cycle analysis. **P<0.01 and ***P<0.001 compared with the miR-145 mimics group. The data are presented as the mean ± standard deviation values.
Abbreviations: ARF6, ADP-ribosylation factor 6; NC, negative control.
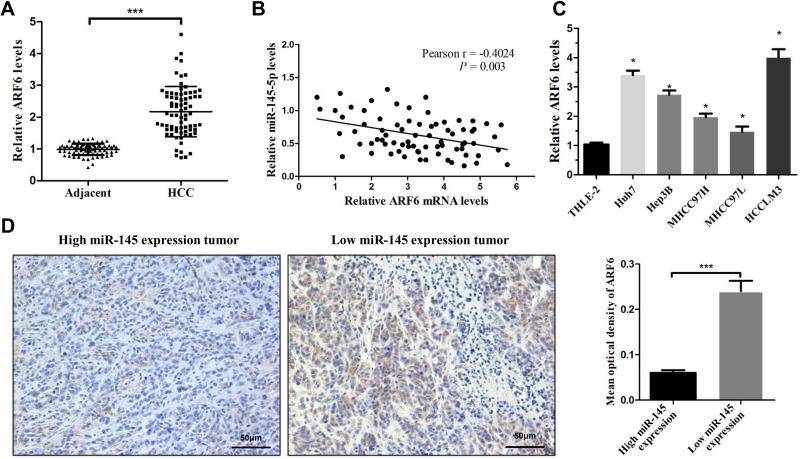
ARF6 is Significantly Overexpressed in HCC and Negatively Correlated with the Expression of miR-145
Due to the metastatic effect of ARF6 in numerous types of tumor, the expression level of ARF6 in HCC tissues was first determined. HCC tissues exhibited significantly higher expression levels of ARF6 compared with ANLTs (Figure 5A). Interestingly, a negative correlation was also found between miR-145 and ARF6 expression (Figure 5B). Subsequently, the expression level of ARF6 in HCC cell lines was further examined. The expression level of ARF6 was confirmed via RT-qPCR to be increased in all HCC cell lines, and was the highest in HCCLM3 and Huh7 cells (Figure 5C). The expression levels of ARF6 in HCC tissues were also quantized with different miR-145 levels by IHC (Figure 5D). Tumors with lower levels of miR-145 tended to overexpress ARF6 and, conversely, tumors that overexpressed miR-145 showed lower levels of ARF6. Collectively, these data suggested that dysregulation of miR-145 was associated with the overexpression of ARF6 in HCC.
Figure 5.
ARF6 was significantly upregulated in HCC tissues and negatively correlated with miR-145 in HCC. (A) Real-time PCR was performed to measure ARF6 mRNA levels in HCC tissues and their matched ANLTs (n=75). ***P<0.001 compared with ANLTs. (B) In HCC tissues, ARF6 levels were negatively correlated with miR-145-5p levels (n=75). (C) ARF6 mRNA levels were measured in HCC cell lines and THLE-2 cells. *P<0.05 compared with THLE-2 cells. (D) Representative immunohistochemical images of ARF6 expression in HCC tissues with different miR-145 levels. ***P<0.001 compared between the two groups. The data are presented as the mean ± standard deviation values.
Abbreviations: HCC, hepatocellular carcinoma; ARF6, ADP-ribosylation factor 6.
Discussion
HCC is a commonly occurring tumor among malignancies worldwide. Due to the fast growth rate and the high recurrence rate of tumors after operation, only 30% of patients with HCC are able to be cured. Despite great efforts to improve treatment measures, the mortality of patients with HCC remains high, seriously threatening human life and health.18 Due to abnormalities in multiple signal transduction pathways and dysregulation of tumor suppressor genes and oncogenes in normal liver cells, these signaling pathways form a complex molecular regulatory network that cooperatively promotes the occurrence and metastasis of HCC.19 Many recent studies have confirmed that a variety of miRNAs can participate in the biological behavior of tumors by regulating the expression of target genes and that they have value in the diagnosis and prognosis of malignant tumors.20 Abnormally expressed miRNAs can act as protooncogenes (eg, miR-222-3p) or as tumor suppressor genes (eg, miR-124a) to influence the development and metastasis of numerous types of malignancy.21,22 A large number of studies have confirmed that miR-145-5p is downregulated in HCC, although its underlying effects on HCC remain unclarified.23 In the present study, it was shown that miR-145 was downregulated not only in HCC tissues but also in several HCC cell lines. The lowest level was observed in the HCCLM3 cell line. Notably, miR-145 levels were negatively correlated with tumor size, portal vein tumor embolus and tumor stage and positively correlated with disease-free survival and overall survival. However, the follow-up time in this study was short; if the follow-up time was to have been extended, the results would have been more convincing. Collectively, these findings showed that miR-145 was associated with HCC metastasis and prognosis. Therefore, it was further confirmed that restoration of miR-145 negatively regulated proliferation, migration, and invasion and induced apoptosis in HCC cells.
ARF6 is localized on the plasma membrane and several endosome membranes in a variety of cells, including nerve cells and liver cells.24,25 ARF6 is involved in regulating plasma membrane transport and intracellular actin assembly;10 therefore, it is involved in diverse physiological functions, including vesicle endocytosis, secretion, neurite outgrowth, cell adhesion, cell migration, and cancer metastasis. This observation indicates that the functional specificity of ARF6 in different types of cells may be caused by different upstream regulatory factors, or differential spatiotemporal expression of their regulatory factors. A previous study reported that the expression of the ARF6 gene was higher in glioblastoma tissues compared with matched adjacent tissues.17 Survival curve analysis proved that ARF6 gene expression was significantly positively correlated with distant metastasis, cancer death, and prognosis, suggesting its involvement in cancer cell metastasis. Taken together, these findings reveal that ARF6 exerts a key role in tumor metastasis. Notably, it was confirmed that ARF6 expression not only was increased in several HCC cell lines but it was also found to be significantly higher in HCC tissues compared with ANLTs. Therefore, ARF6 may play an important role in HCC metastasis.
Hsu et al demonstrated that miR-145 suppressed cell migration and invasion in upper tract urothelial carcinoma by targeting ARF6.26 miR-145 was also reported to regulate hepatocellular carcinoma progression and proliferation of epithelial ovarian cancer by targeting SPATS2 and SMAD4.27,28 In order to reveal the relationship between miR-145 and ARF6 in HCC, some further experiments were implemented. ARF6 was subsequently predicted to be a downstream target gene of miR-145 by four different miRNA targeting prediction tools, and the targeting relationship was confirmed through dual-luciferase reporter assay. Interestingly, the present data revealed that miR-145 was negatively correlated with ARF6 expression in HCC tissues. Tumors with lower levels of miR-145 tended to overexpress ARF6 and, conversely, tumors that overexpressed miR-145 showed lower levels of ARF6. Furthermore, restoration of ARF6 rescued miR-145-inhibited tumor migration and invasion. Collectively, these findings showed that miR-145 inhibits tumor metastasis by targeting ARF6.
In summary, the present study has confirmed miR-145-5p downregulation in HCC, and that this is associated with poor prognosis. miR-145-5p inhibits invasion and metastasis in vitro by directly targeting ARF6, indicating that miR-145-5p and ARF6 could be novel targets for preventing HCC metastasis.
Funding Statement
The present study was supported by grants from Primary Research and Development Plan of Zhejiang Province (2015C33203), and National Key Research and Development Program of China (2016YFC1101504).
Ethics Approval
All tissues used in this study were acquired with written informed consent. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All human experiments were approved by the Clinical Research Ethics Committee of the First Affiliated Hospital, Zhejiang University School of Medicine (Hangzhou, China).
Disclosure
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi: 10.1016/j.jhep.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 3.Fang JH, Zhou HC, Zhang C, et al. A novel vascular pattern promotes metastasis of hepatocellular carcinoma in an epithelial-mesenchymal transition-independent manner. Hepatology. 2015;62(2):452–465. doi: 10.1002/hep.27760 [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 6.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707 [DOI] [PubMed] [Google Scholar]
- 7.Ishii H, Vodnala SK, Achyut BR, et al. miR-130a and miR-145 reprogram Gr-1CD11b myeloid cells and inhibit tumor metastasis through improved host immunity. Nat Commun. 2018;9(1):2611. doi: 10.1038/s41467-018-05023-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong QW, Lung RW, Law PT, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135(1):257–269. doi: 10.1053/j.gastro.2008.04.003 [DOI] [PubMed] [Google Scholar]
- 9.Gai X, Tang B, Liu F, et al. mTOR/miR-145-regulated exosomal GOLM1 promotes hepatocellular carcinoma through augmented GSK-3β/MMPs. J Genet Genomics. 2019;46(5):235–245. doi: 10.1016/j.jgg.2019.03.013 [DOI] [PubMed] [Google Scholar]
- 10.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7(5):347–358. doi: 10.1038/nrm1910 [DOI] [PubMed] [Google Scholar]
- 11.Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem. 2003;278(43):41573–41576. doi: 10.1074/jbc.R300026200 [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto S, Furukawa S, Hashimoto A, et al. KRASARF6 and AMAP1 are major targets of and mutations to promote invasion, PD-L1 dynamics, and immune evasion of pancreatic cancer. Proc Natl Acad Sci USA. 2019;116(35):17450–17459. doi: 10.1073/pnas.1901765116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao W, Zhang C, Li W, et al. Promoter Methylation-Regulated miR-145-5p Inhibits Laryngeal Squamous Cell Carcinoma Progression by Targeting FSCN1. Mol Ther. 2019;27(2):365–379. doi: 10.1016/j.ymthe.2018.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi S, Su L, Li J, et al. Arf6-driven endocytic recycling of CD147 determines HCC malignant phenotypes. J Exp Clin Cancer Res. 2019;38(1):471. doi: 10.1186/s13046-019-1464-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onodera Y, Nam JM, Hashimoto A, et al. Rab5c promotes AMAP1-PRKD2 complex formation to enhance β1 integrin recycling in EGF-induced cancer invasion. J Cell Biol. 2012;197(7):983–996. doi: 10.1083/jcb.201201065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato H, Hatanaka KC, Hatanaka Y, et al. High level expression of AMAP1 protein correlates with poor prognosis and survival after surgery of head and neck squamous cell carcinoma patients. Cell Commun Signal. 2014;12(1):17. doi: 10.1186/1478-811X-12-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Wang J, Ng SS, et al. Adenosine diphosphate-ribosylation factor 6 is required for epidermal growth factor-induced glioblastoma cell proliferation. Cancer. 2009;115(21):4959–4972. doi: 10.1002/cncr.24550 [DOI] [PubMed] [Google Scholar]
- 18.Forner A, Reig M, Bruix J. Reig M and Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi: 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 19.Lu M, Zhu WW, Wang X, et al. ACOT12-Dependent Alteration of Acetyl-CoA Drives Hepatocellular Carcinoma Metastasis by Epigenetic Induction of Epithelial-Mesenchymal Transition. Cell Metab. 2019;29(4):886–900. doi: 10.1016/j.cmet.2018.12.019 [DOI] [PubMed] [Google Scholar]
- 20.Lupini L, Pepe F, Ferracin M, et al. Over-expression of the miR-483-3p Overcomes the miR-145/TP53 Pro-apoptotic Loop in Hepatocellular Carcinoma. Oncotarget. 2016;7(21):31361–31371. doi: 10.18632/oncotarget.8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei F, Ma C, Zhou T, et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol cancer. 2017;16(1):132. doi: 10.1186/s12943-017-0694-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu L, Wang S, Lin X, Lu Y, Gu P. MicroRNA-124a inhibits cell proliferation and migration in liver cancer by regulating interleukin-11. Mol Med Rep. 2018;17(3):3972–3978. doi: 10.3892/mmr.2017.8348 [DOI] [PubMed] [Google Scholar]
- 23.Wang B, Dong W, Li X. miR-145-5p Acts as a Novel Tumor Suppressor in Hepatocellular Carcinoma Through Targeting RAB18. Technol Cancer Res Treat. 2019;18:1533033819850189. doi: 10.1177/1533033819850189 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Hernández-Deviez DJ, Casanova JE, Wilson JM. Regulation of dendritic development by the ARF exchange factor ARNO. Nat Neurosci. 2002;5(7):623–624. doi: 10.1038/nn865 [DOI] [PubMed] [Google Scholar]
- 25.Wu B, Wang Y, Yang XM, et al. Basigin-mediated redistribution of CD98 promotes cell spreading and tumorigenicity in hepatocellular carcinoma. J Exp Clin Cancer Res. 2015;34:110. doi: 10.1186/s13046-015-0226-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu WC, Li WM, Lee YC, et al. MicroRNA-145 Suppresses Cell Migration and Invasion in Upper Tract Urothelial Carcinoma by Targeting ARF6. FASEB J. 2020;34(4):5975–5992. doi: 10.1096/fj.201902555R [DOI] [PubMed] [Google Scholar]
- 27.Dong G, Zhang S, Shen S, et al. SPATS2, negatively regulated by miR-145-5p, promotes hepatocellular carcinoma progression through regulating cell cycle. Cell Death Dis. 2020;11(10):837. doi: 10.1038/s41419-020-03039-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Zhang X, Li W, Chen Y. MicroRNA-145-5p regulates the proliferation of epithelial ovarian cancer cells via targeting SMAD4. J Ovarian Res. 2020;13(1):54. doi: 10.1186/s13048-020-00656-1 [DOI] [PMC free article] [PubMed] [Google Scholar]