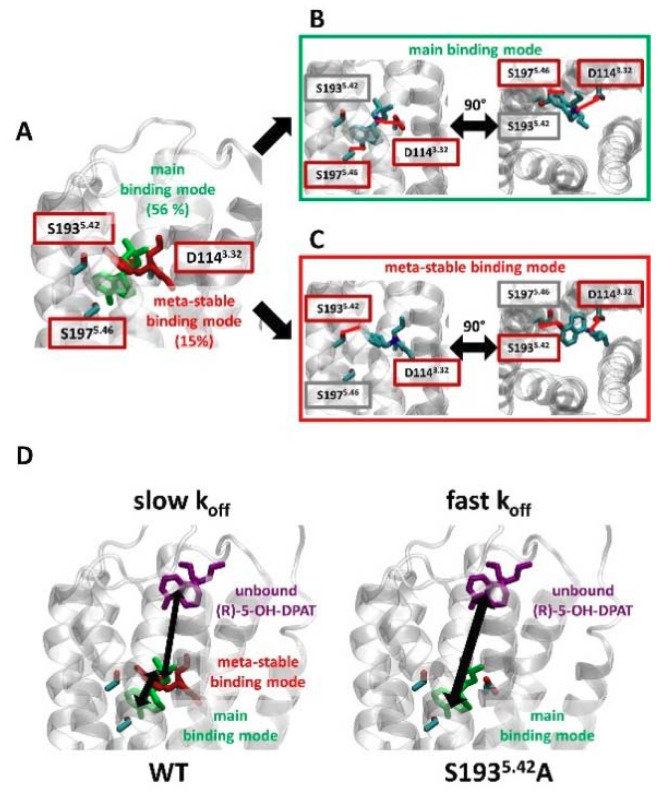
Figure 5.
(R)-5-OH-DPAT forms a meta-stable binding mode with S1935.42. (A) Clustering simulations of (R)-5-OH-DPAT in complex with WT D2R, based on the RMSD of the ligand, reveal two binding modes. The main binding mode was maintained over 56% of the simulation frames (green) and a meta-stable binding mode was maintained over 15% of the simulation frames (red). (B,C). For each of the binding modes, the studied position S1935.42, as well as residues that form polar interactions with the ligand, are shown. These polar interactions are highlighted with red lines. (D) A model explaining the slow Koff values observed for the unbinding of (R)-5-OH-DPAT from WT D2R. Before dissociating from the receptor (purple conformation), (R)-5-OH-DPAT bound in the main binding mode (green) assumes a meta-stable binding mode (red). When bound in this meta-stable binding mode, (R)-5-OH-DPAT can revert to the main binding mode or proceed to an unbound conformation. Hence, the dissociation of (R)-5-OH-DPAT is effectively slowed down by S1935.42. In comparison, at the S1935.42A mutant receptor, the absence of the meta-stable binding mode permits fast exchange between the bound and unbound conformations of (R)-5-OH-DPAT.