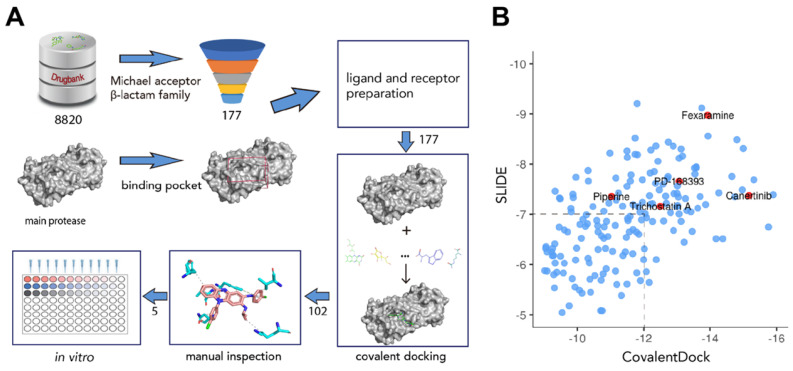
Figure 1.
Overview of the structure-based covalent docking virtual screening workflow. (A) Small molecules in DrugBank database (8,820 compounds) were pre-filtered for covalent docking (Michael acceptor and β-lactam family). Potential binding pockets were predicted for SARS-CoV-2 Mpro. Then each compound (177 candidates) was docked against binding pocket with CovalentDock. Binding poses (102 compounds) were manually inspected for downstream experimental validation based on certain criteria including relative binding affinity, ligand efficiency, hydrogen bond and hydrophobic contacts etc. (B) Scatter plot showing the distribution of CovalentDock and SLIDE scores of 177 docked poses. Five selected compounds were highlighted in red.