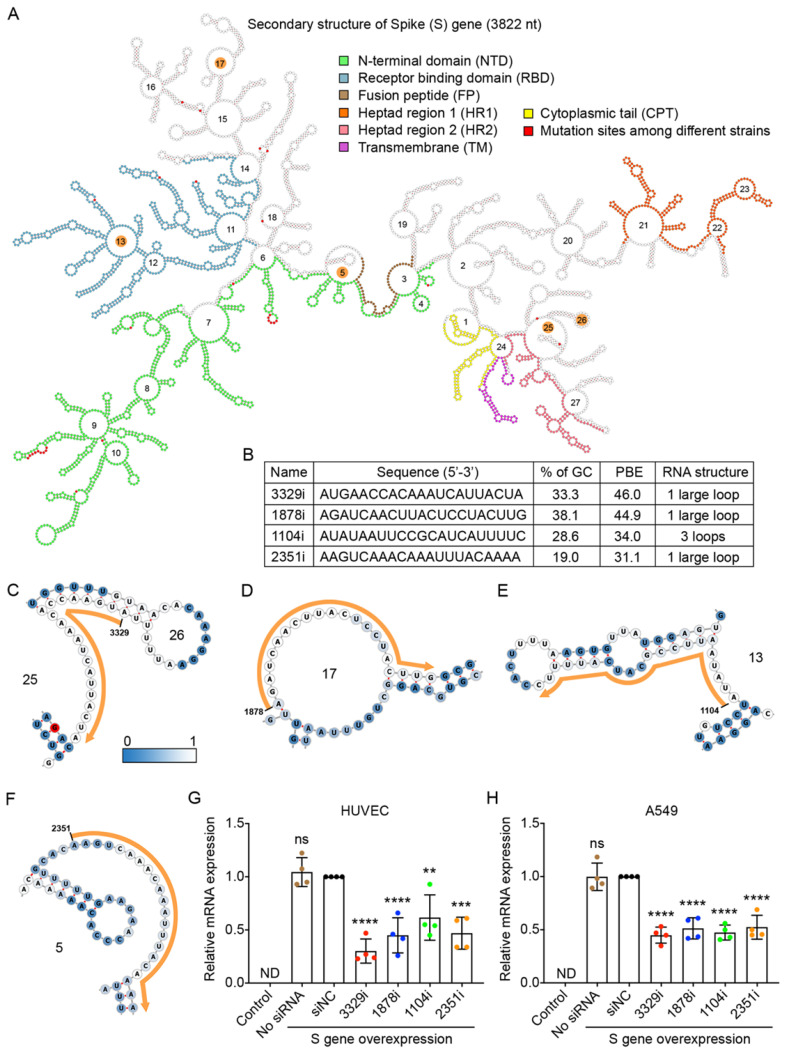
Figure 3.
Selection of high PBE siRNAs for the spike (S) gene. A. The secondary structure of the RNA molecule of the S gene. The nucleotide sequences corresponding to functional protein domains are labeled in different colors. Mutated sites among the nine viral strains are labeled in red. The loops numbered 5, 13, 17, 25 and 26 are highlighted in orange, as these regions were selected for siRNA targeting. B. The sequence, GC content, PBE and RNA structure of the four selected siRNAs. C-F. Detailed structures of targeted regions by four siRNAs, 3329i (C), 1878i (D), 1104i (E) and 2351i (F), on the S gene with the siRNA sequence outlined in orange. The color of each nucleotide reflects the possibility of being single-stranded. G-H. Real-time qPCR was used to measure the relative expression of the S gene in both HUVECs (G) and A549 cells (H) after transfection with the plasmid DNA of the S gene and individual siRNAs at 24 hours. Cells that were not transfected were used as a background control. All values were normalized to the siNC group, in which cells were transfected with the S gene and a negative control siRNA simultaneously. In No siRNA group, only plasmid DNA was transfected.