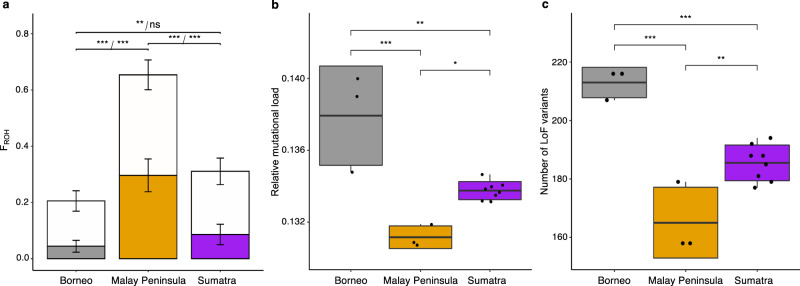
Fig. 3. Inbreeding, relative mutational load and number of loss-of-function (LoF) variants in three modern populations of Sumatran rhinoceros (Dicerorhinus sumatrensis).
a Inbreeding estimated as the average proportion of the genome in runs of homozygosity (FROH). Open bars show total proportion of the genome in ROH ≥ 100 kb and solid bars show proportions in ROH of length ≥ 2 Mb. Bars extending from the mean values represent the standard deviation (two-sided pairwise t-test, FROH ≥ 100 kb: pBorneo-MalayP = 4.2e−07, pBorneo-Sumatra = 0.0066, pMalayP-Sumatra = 6.4e−07, FROH ≥ 2 Mb: pBorneo-MalayP = 2.2e−05, pBorneo-Sumatra = 0.15, pMalayP-Sumatra = 2.2e−05). b Mutational load estimated with GERP scores. Individual relative mutational load was measured as the sum of all derived alleles multiplied by their conservation-score over the total number of derived alleles (see ‘Methods’). Only derived alleles above conservation-score of 1 (i.e., non-neutral) were included (two-sided pairwise t-test, pBorneo-MalayP = 0.00013, pBorneo-Sumatra = 0.001, pMalayP-Sumatra = 0.012. c Number of LoF variants using an annotation of 33,026 gene predictions for a white rhinoceros genome assembly (see ‘Methods’) (two-sided pairwise t-test, pBorneo-MalayP = 2.2e−05, pBorneo-Sumatra = 0.0004, pMalayP-Sumatra = 0.0019). Middle thick lines within boxplots and bounds of boxes represent mean and standard deviation, respectively. Vertical lines represent minima and maxima. n = 14, ***p < 0.001, **p < 0.01, *p < 0.05, ns = non-significant, p-values were not adjusted for multiple comparisons.